Topology Independent Comparison of Biomolecular 3D Structures
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
In our effort to unearth cases of DNA-protein complexes, we have detected instances of DNA-protein binding sites that look geometrically similar even when the proteins belong to different fold families. An example would be the CLICK search for the transcription factor GCN4 (PDB code 1YSA and SCOP entry: h.1.3.1). The DNA-protein binding regions of GCN4 are DNA chains A and B, and protein chain C from residue 228 to 249 and chain D from residue 230 to 250 (figure 1).
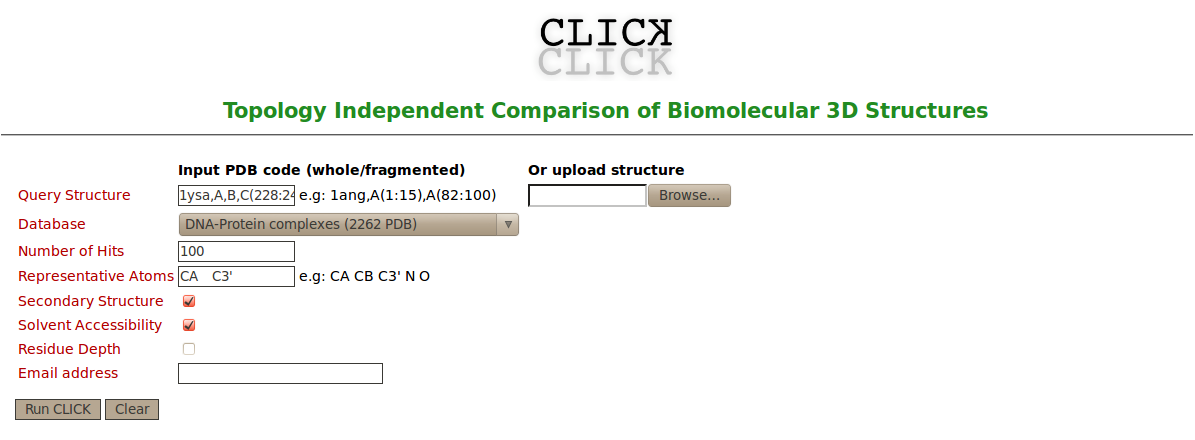
To the best of our knowledge CLICK is the only structure comparison server that can handle multiple types of biomolecular structures independent of topology and even deal with fragments or complexes of these molecules. Using the structure of DNA-protein binding regions of GCN4 as query, we searched the PDB for topologically different regions of DNA-protein complexes that matched these binding sites structurally (figure 2A). In this search, C3' and Cα are used as representative atoms for DNA-protein complexes. Searching over a dataset of 2262 representatives of DNA-protein complexes, 11 hits were recovered with a topologically different and Z-score above 2.5 (figure 2B). These hits are 2WTY (the 3rd hit and there is no SCOP id for this hit, figure 3), 3A5T (the 4th hit and No SCOP id, figure 4), 2WT7 (the 7th hit and No SCOP id, figure 5), 1AM9 (the 10th hit and SCOP entry: a.38.1.1, figure 6), 3VEB (the 22th hit and No SCOP id, figure 7), 3VEA (the 28th hit and No SCOP id, figure 8), 2QL2 (the 29th hit and No SCOP id, figure 9), 4ATI (the 30th hit and No SCOP id, figure 10), 1MDY (the 10th hit and SCOP entry: a.38.1.1, figure 11), 4D8J (the 34th hit and No SCOP id, figure 12), 4H10 (the 35th hit and No SCOP id, figure 13). The superimpositions of these 11 hits and the DNA-protein binding regions of GCN4 show that CLICK produced accurate alignments of DNA-protein interactions (figures 3-13). These superimpositions with the diverse proteins help determine the common factors in DNA recognition. Though protein regions of these 11 hits shown in this example are not related to the protein region of the transcription factor of GCN4 in sequence or structure, the Arginine residues (shown in red in the alignments and in stick representation in the figures 3-13) that mediate the protein-DNA interaction are conserved in their alignments and in almost identical spatial locations. These results suggest CLICK is a powerful computational tool to determine such similarities and could be used in functional annotation and molecular design. |

|
| Figure 1. The DNA-protein binding regions of the transcription factor of GCN4 (PDB code 1YSA and SCOP entry: h.1.3.1) include DNA chains A and B, and protein chain C from residue 228 to 249 and chain D from residue 230 to 250. |
|
| Figure 2A. Snapshot of the CLICK web server of input page for searching similar structures to the DNA-protein binding regions of GCN4 over a dataset of 2262 representatives of DNA-protein complexes |
| [back to top] |
|
|

|
|
Figure 2B. Snapshot of the CLICK web server of output page (http://mspc.bii.a-star.edu.sg/minhn/output_db/142355945679.html) for searching similar structures to the DNA-protein binding regions of GCN4 over a dataset of 2262 representatives of DNA-protein complexes |
| [back to top] |
|
|
|

|
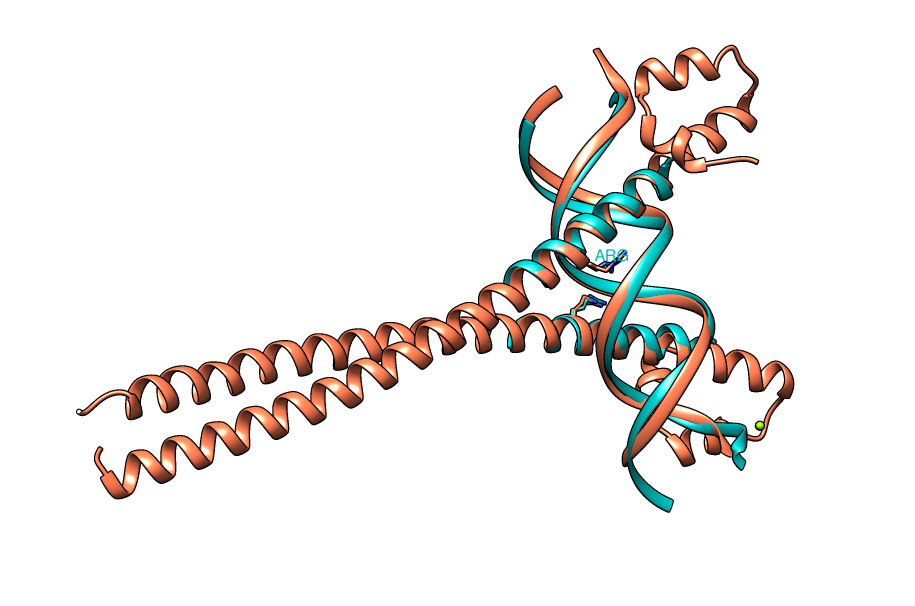
| Figure 3. CLICK superposition of transcription factor MafB (pdb code 2WTY, the 3rd hit and salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an structure overlap (SO) of 90.36% and RMSD of 1.40Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/142355447563.html). There are two Arginine residues (shown in red in the alignment and in stick representation in the figure) that mediate the protein-DNA interaction are conserved in the alignment and in identical spatial locations. |
| [back to top] |
|
|
|
|
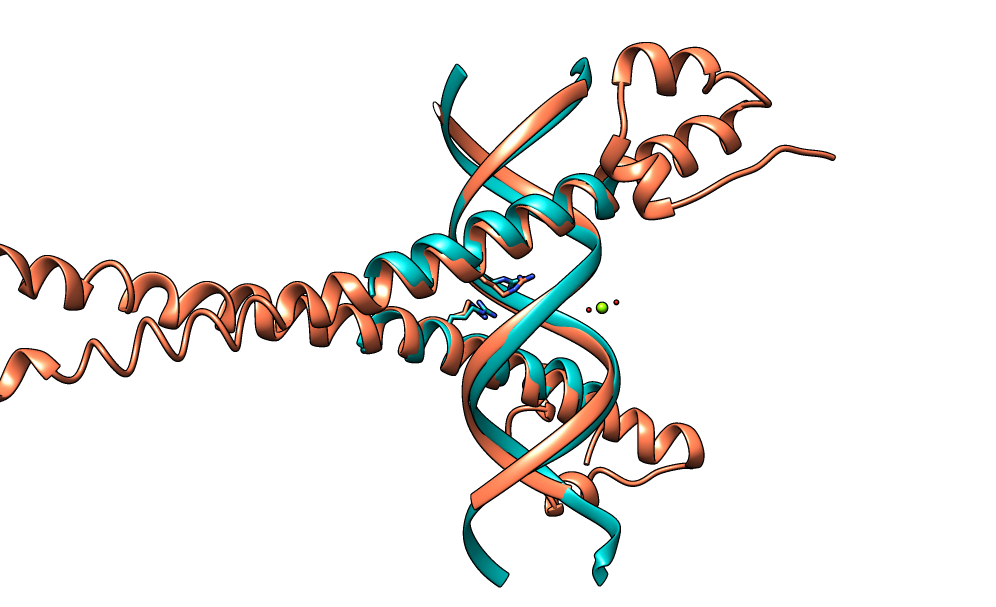
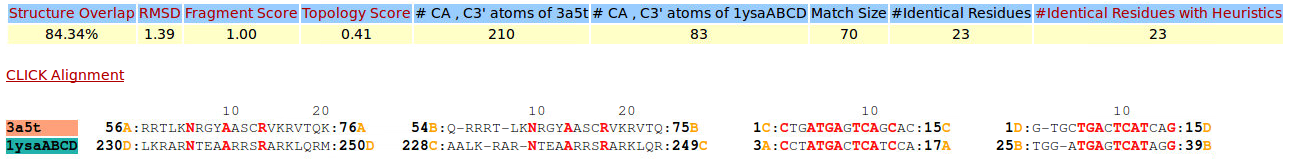
| Figure 4. CLICK superposition of 3A5T (the 4th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 84.34% and RMSD of 1.39Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/142373323247.html). There are two Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|
|

|
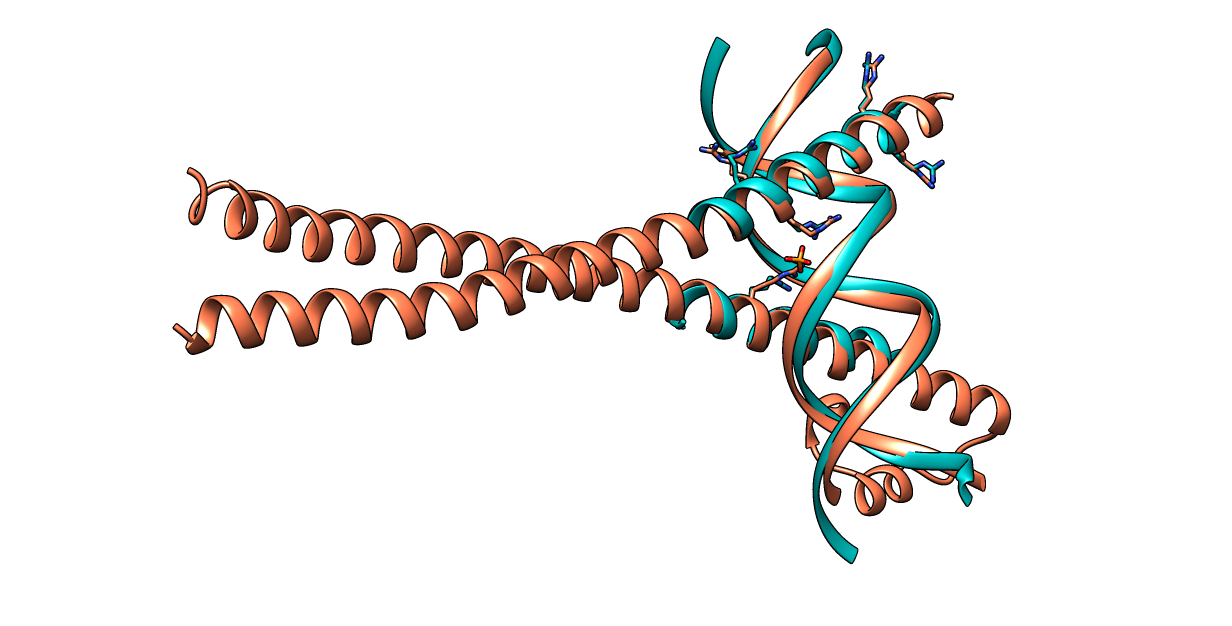
| Figure 5. CLICK superposition of 2WT7 (the 7th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 85.54% and RMSD of 1.51Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/142380326831.html). There are five Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|
|

|
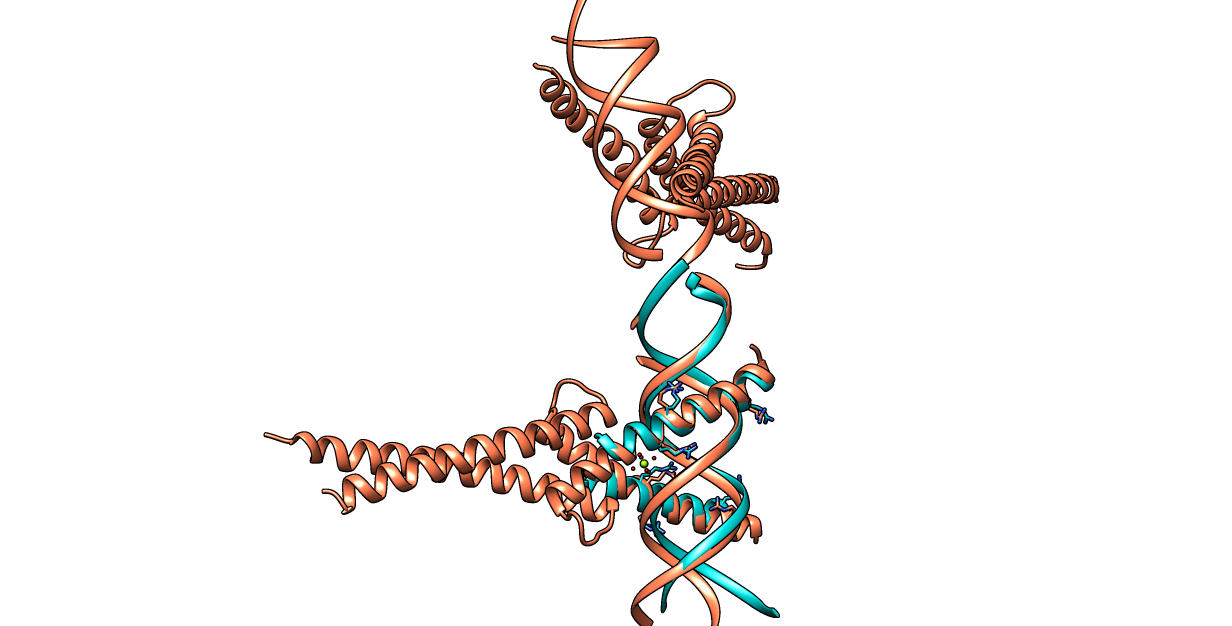
| Figure 6. CLICK superposition of 1AM9 (the 10th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 91.57% and RMSD of 1.78Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/142380345957.html). There are five Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|
|
|
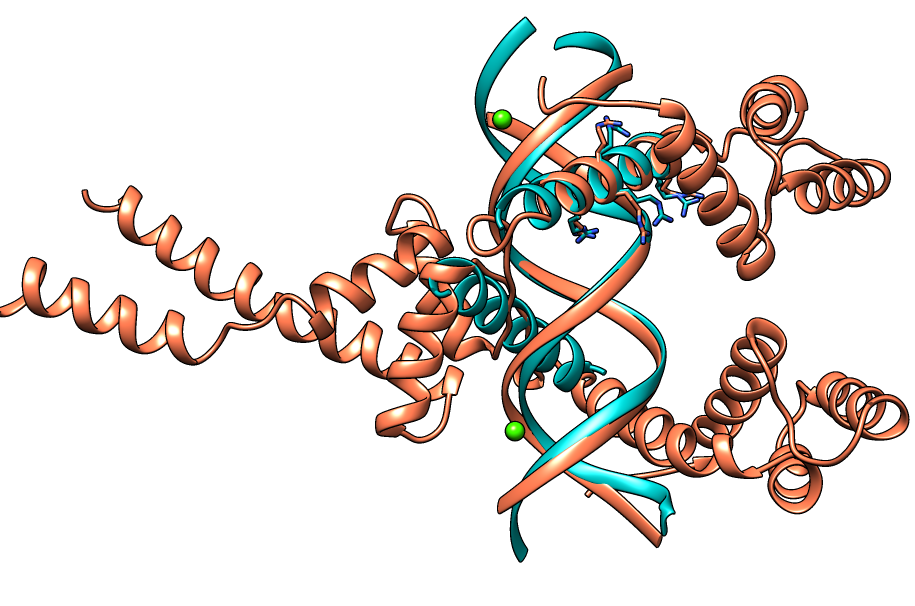
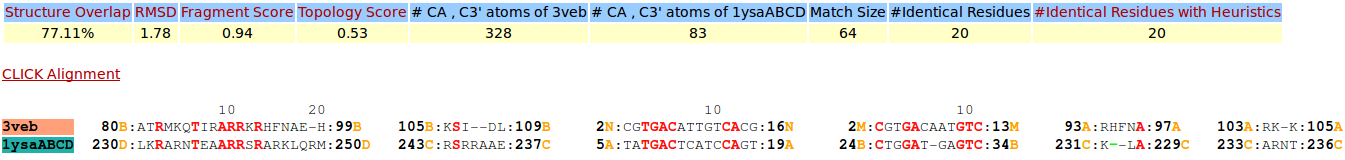
| Figure 7. CLICK superposition of 3VEB (the 22th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 77.11% and RMSD of 1.78Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/14238035398.html). There are four Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|
|

|
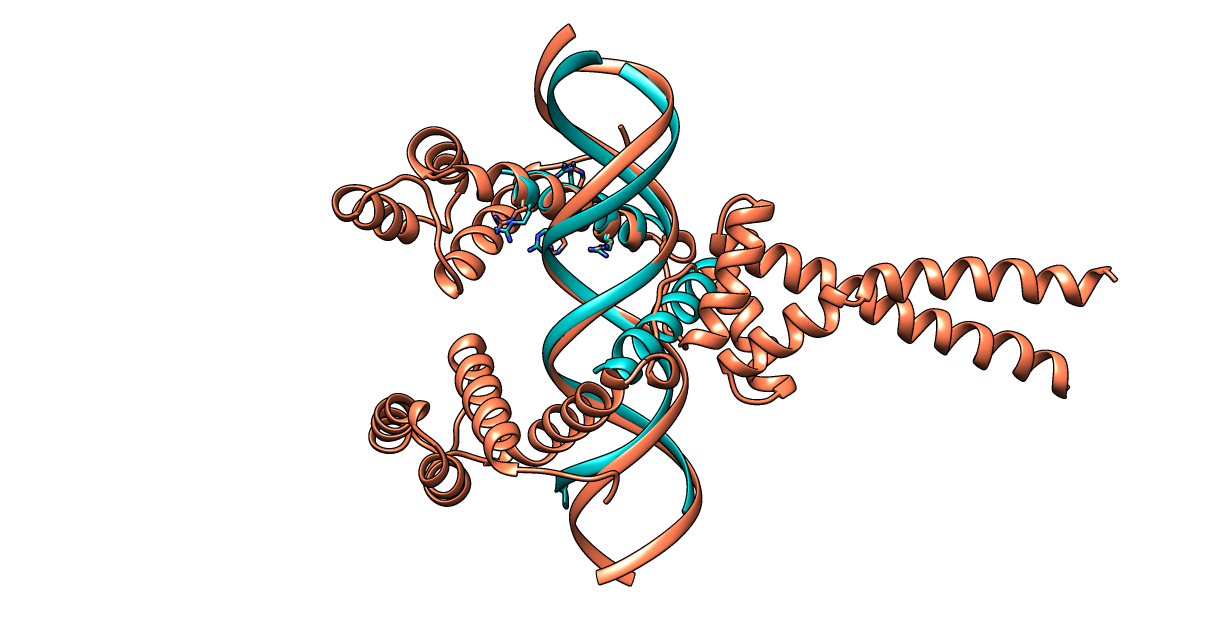
| Figure 8. CLICK superposition of 3VEA (the 28th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 75.90% and RMSD of 1.81Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/142380365351.html). There are four Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|
|
|
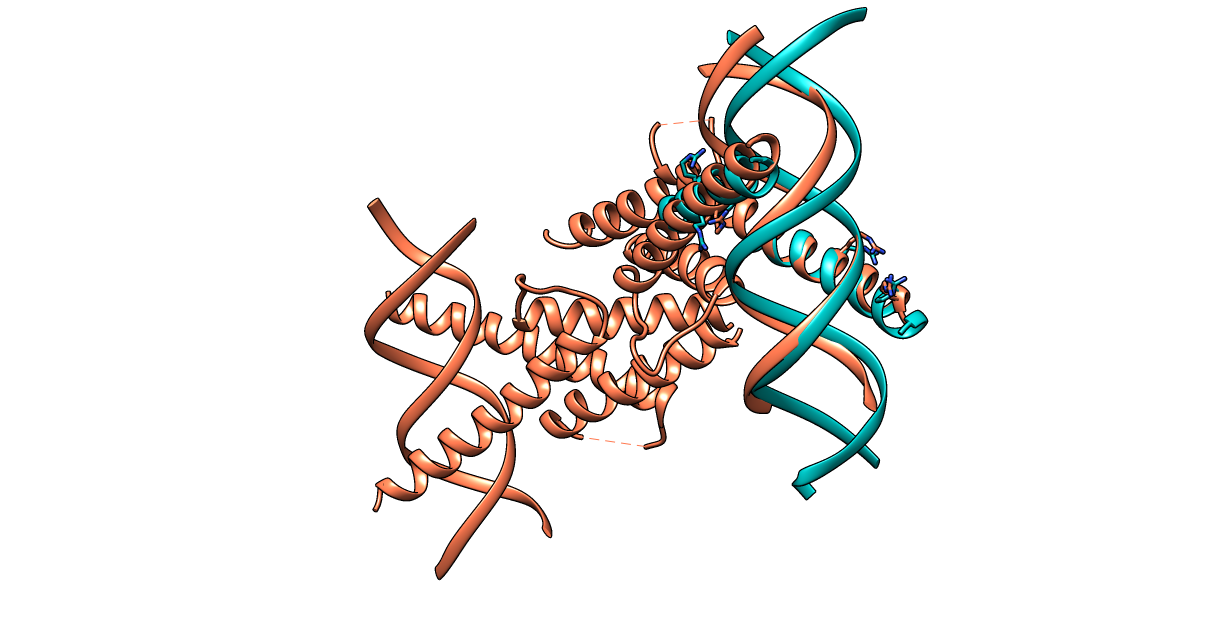
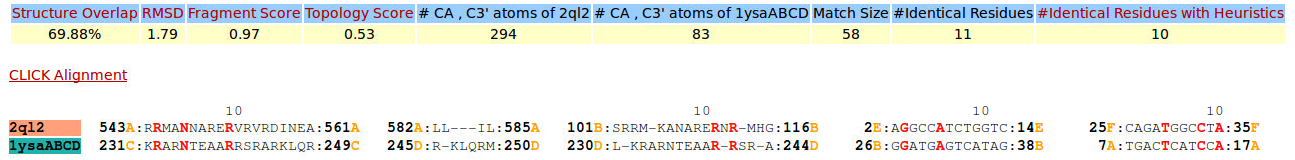
| Figure 9. CLICK superposition of 2QL2 (the 29th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 69.88% and RMSD of 1.79Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/142380383038.html). There are four Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|

|
|
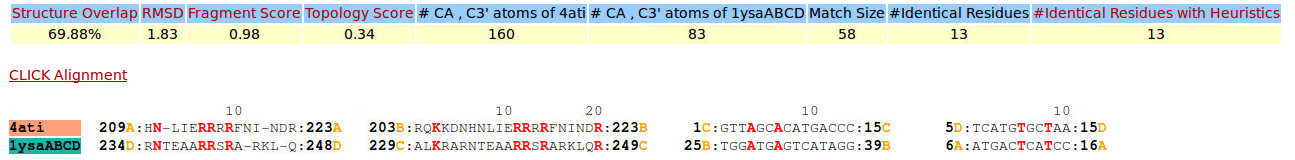
| Figure 10. CLICK superposition of 4ATI (the 30th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 69.88% and RMSD of 1.83Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/142380389734.html). There are seven Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|

|
|
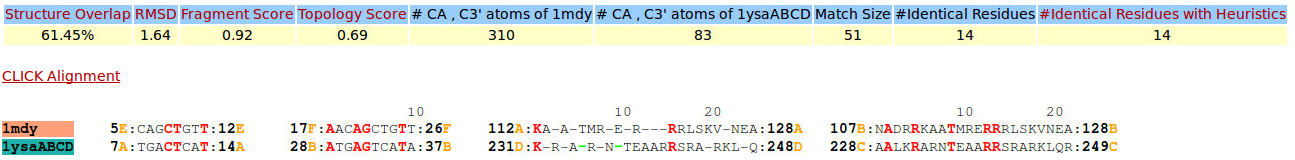
| Figure 11. CLICK superposition of 1MDY (the 31th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 61.45% and RMSD of 1.64Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/142380402539.html). There are four Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|

|
|
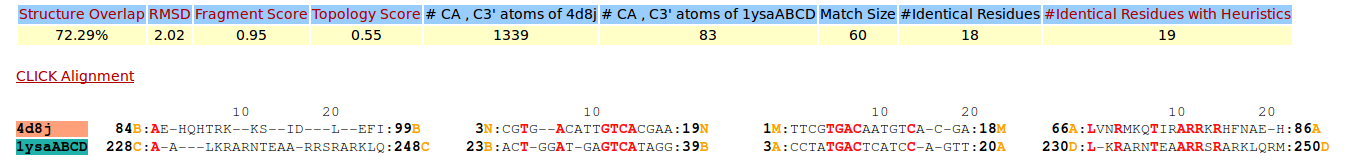
| Figure 12. CLICK superposition of 4D8J (the 34th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 72.29% and RMSD of 2.02Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/14238043179.html). There are four Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
| [back to top] |
|
|
|
|
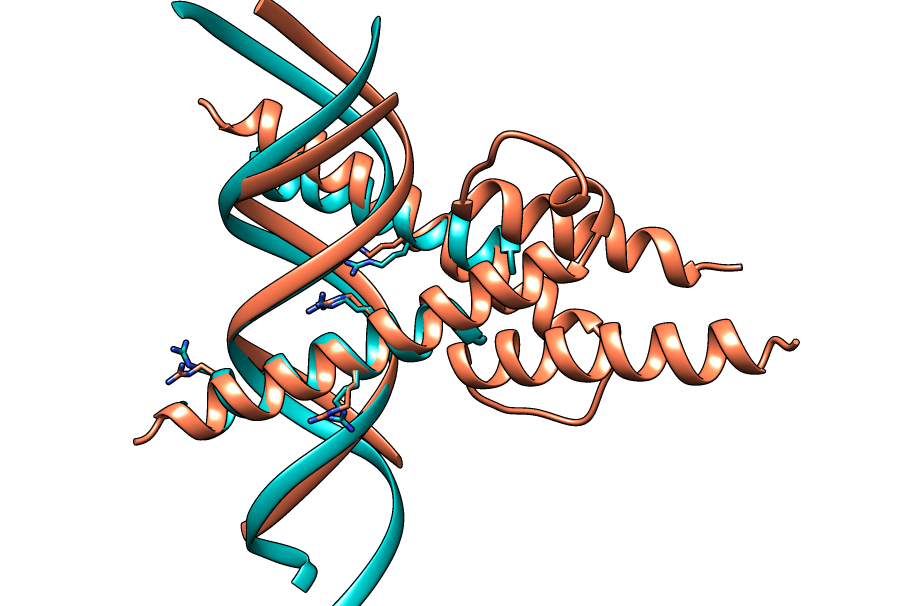
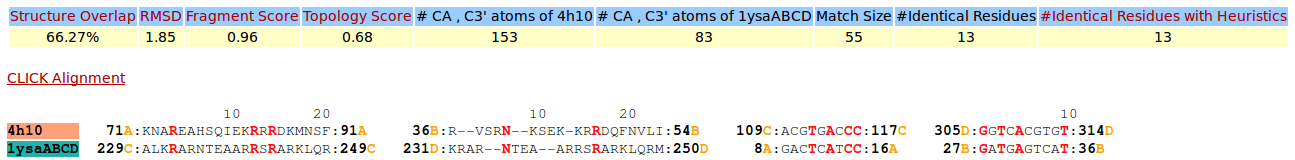
| Figure 13. CLICK superposition of 4H10 (the 35th hit, salmon color) and the DNA-protein binding regions of GCN4 (green) using representative C3' and Cα atoms with an SO of 66.27% and RMSD of 1.85Å (http://mspc.bii.a-star.edu.sg/minhn/test_data/14238062037.html). There are four Arginine residues of the protein-DNA interaction that are conserved in the alignment and in almost identical spatial locations. |
|
|
| Pictures rendered using Chimera (http://www.cgl.ucsf.edu/chimera/) |
| [back to top] |