|
|
|
|
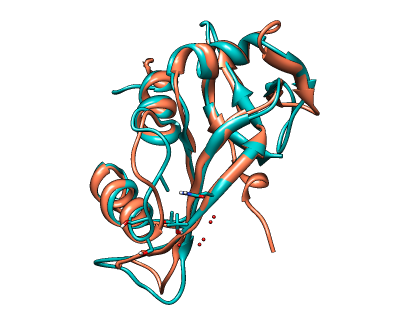
Picture rendered using Chimera (http://www.cgl.ucsf.edu/chimera/)
|
|
|

|
|
CLICK superposition of Human Angiogenin, 1ang (salmon) and RNAseA, 7rsa (green) with an SO of 84.55%, RMSD of 1.24Å, and fragment and topology scores of 0.99 and 1.00 respectively.
|
|
|
|
Picture rendered using Chimera (http://www.cgl.ucsf.edu/chimera/)
|
|
|

|
|
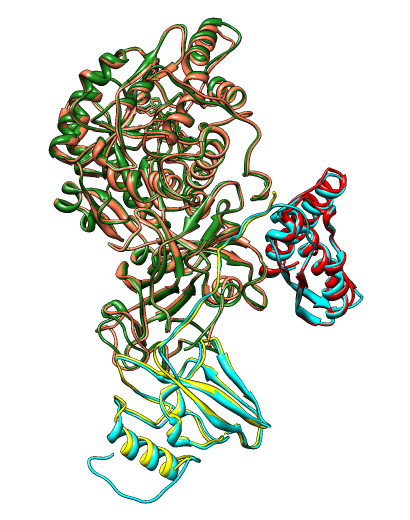
An alignment between 2 PDB files (1ubp chains A (red), B (yellow), and
C (salmon); and 1e9y chains A (cyan) and B (forest green)) of proteins with multiple chains.
Chains A and B of 1ubp superimpose onto chain A of 1e9y, and chain C of
1ubp superimpose onto chain B of 1e9y.
|
|
| [back to top] |
|
|
|
Picture rendered using Chimera (http://www.cgl.ucsf.edu/chimera/)
|
|
|

|
|
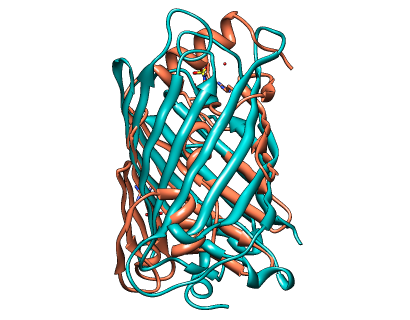
CLICK superposition with different SCOP class of 1iwm:A (salmon & SCOP entry: b.125.1.2) and 1oxe:A (green & SCOP entry: d.22.1.1) with an SO of 63.28%, RMSD of 2.27Å, and fragment and topology scores of 0.62 and 0.57 respectively.
|
|
| [back to top] |
|
The first alignment of 1aiv and 1ovt
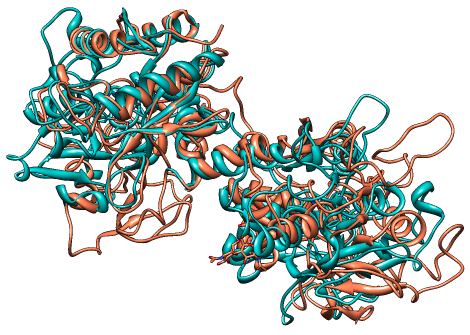
|
|
Picture rendered using Chimera (http://www.cgl.ucsf.edu/chimera/)
|
|
|

|
The second alignment of 1aiv and 1ovt
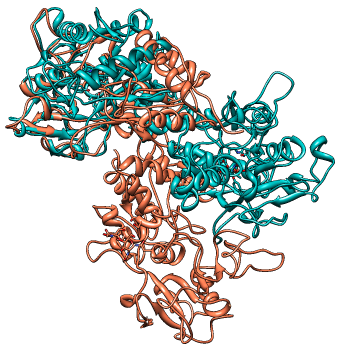
|
|
Picture rendered using Chimera (http://www.cgl.ucsf.edu/chimera/)
|
|
|

|
The third alignment of 1aiv and 1ovt
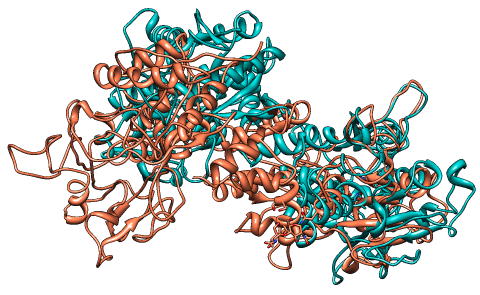
|
|
Picture rendered using Chimera (http://www.cgl.ucsf.edu/chimera/)
|
|
|
10 20 30 40
1aiv 492A:SPPN-SR-LCQLCQGSGG-IPPEKCVASS-HEK-YF-GYTG-A-L-RC:530A
1ovt 492A:SPPNSR-L-CQLCQGS-GGIPPEKCVA-SSHEKYF-G--Y-T-G-A-L:528A |
|
Individual domains in
1aiv (salmon) have a structural equivalence in 1ovt (green), but
the relative orientations between these domains differ in the two
structures.
|
|
| [back to top] |
|
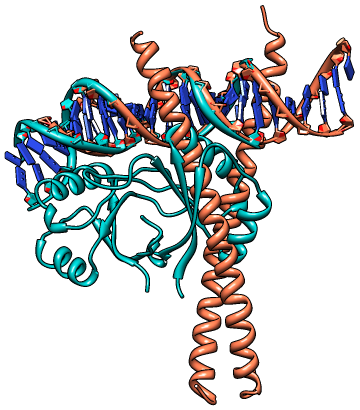
|
|
Picture rendered using Chimera (http://www.cgl.ucsf.edu/chimera/)
|
|
|
10 10
1ysa 7A:TGACTCATCCAGT-T:20A 21B:AA-ACTGGATGAGTCA:35B
2ayg 1C:GCAACCGAATTCGGT:15C 5D:C-C-GAATTCGGTTGC:18D |
|
CLICK superposition with DNA double helical structures 1ysa (salmon) and
2ayg (green) using representative C3' atom with an SO of 75.00% and RMSD of 1.80Å.
|
|
| [back to top] |