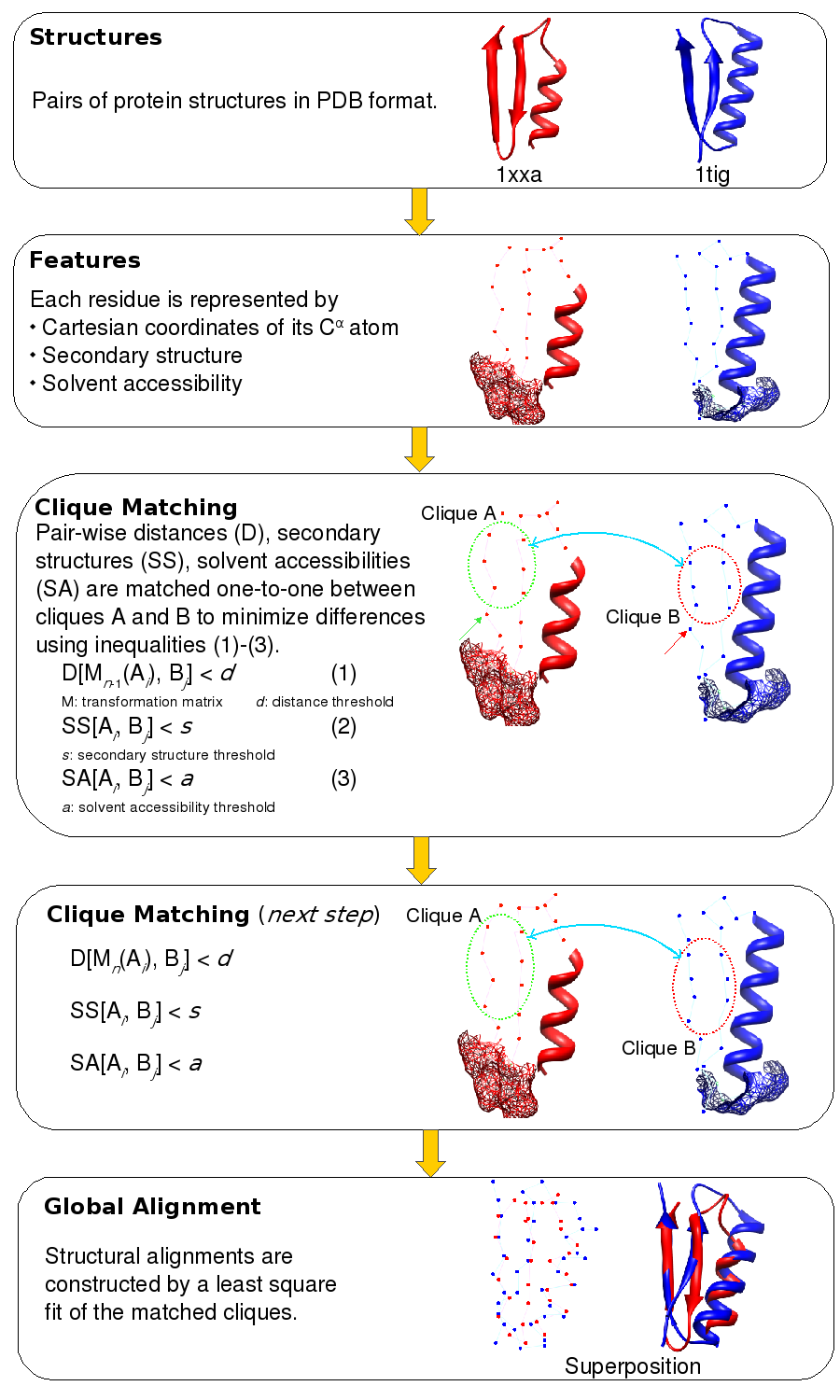
A pictorial representation of the matching of n-body cliques. The structures of two proteins, PDB codes 1xxa chain A and 1tig chain A, are shown in red and blue respectively. The representation of the 3D structure is a composite of the Cα trace, secondary structure, and accessible surface area (shown in mesh representation). The two sets of 6 points encircled in red and blue represent 6-body (n=6) cliques in Clique Matching step. The line that connects them symbolizes the matching of the Cartesian coordinates (by least square fitting), the secondary structure, and the solvent accessible surface area.
A pair of n-body cliques is matched if their RMSD on superimposition is smaller than a preset threshold. RMSD between cliques is calculated by 3D least squares fit. Then all matched pairs of n-body cliques are extended to all possible higher order cliques. To begin with, all possible 3-body cliques are compared to one another. Cliques are then extended to a maximum of 7 constituent residues.
Matching cliques helps in identifying structurally equivalent residues in the two structures. Using these equivalences, a final 3D least squares fit is performed to superimpose the two structures (Global Alignment step). Given that the matching of cliques is not unique, ie. many cliques comparisons could fit the criteria for a match. Of all the possible least square fits, the comparison that yields the best structure overlap is considered.
|