

The Interfacial Character of Antibody Paratopes: Analysis of Antibody-Antigen Structures
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
The paratope of an antibody, which refers in general to the Complementarity Determining Regions (CDRs), is the most important molecular motif for recognition of the epitope of its antigen with specificity and high affinity. In our study, computational methods have been applied to investigate the general properties of contact residues of antibody structures from a non-redundant dataset of 403 antibody-antigen complexes, the largest available dataset used for such studies to date. In addition, previous studies have largely focused on analysis at the residue level while we have delved deeper and dissected out hydrogen bonds, hydrophobic, van der Waals, and ionic interactions. Our results confirm the general principles that have been gleaned from earlier studies based on smaller datasets, and in addition, provide new insights.
From an analysis of the 8603 contact residues of antibodies that we extracted from this dataset, we have confirmed Tyr to be the contact residue with the highest frequency of participation in epitope binding. Our analysis reveals that Tyr is able to interact with epitopes of structurally diverse antigens as it engages in the highest number of hydrogen bonds, hydrophobic, and van der Waals interactions. We have further found that hydrogen bonds, van der Waals, hydrophobic, and ionic interactions made by Tyr, Trp, Ser, Asn, Asp, Thr, Arg, Gly contribute substantially to the interactions between antibody and antigen. Our study confirms His is over-represented in contact residues of antibodies. Furthermore, antibody-antigen interactions can also be mediated by water molecules, which have been known to be important for mediating protein-protein interactions via hydrogen bonds. Yokota et al. have demonstrated the importance of water mediated hydrogen bonds in the interactions between HEL (hen egg white lysozyme) and its HyHEL-10 Fv antibody . However, to the best of our knowledge, there is no reported comprehensive analysis for a large number of structured waters that engage in higher ordered structures at the antibody-antigen interface. From the non-redundant dataset of 403 antibody-antigen complexes, we have found the presence of interfacial waters in 242 complexes. We present evidence that suggests a compelling role of these interfacial waters in interactions of antibodies with a range of antigens differing in shape complementarity. An analysis of our dataset reveals that water molecules mediate a significant number of interactions between antibodies and antigens. Having mined the contact residues from the heavy and light chains of antibody structures with their interfacial water molecules from 403 antibody-antigen complexes, we spatially cluster them using our 3D structure comparison method CLICK. The resultant 368 different clusters are the basis for a new database. Our study shows that structural motifs of contact residues of antibody structures can be identified from this clustering database. Recently, the SAbDab server provides a clustering of antibody CDR conformations . Adolf-Bryfogle et al. have also constructed a database (PyIgClassify) for classification of CDR conformations of antibodies . However, currently there is no database of structural classification for contact residues of antibodies. The results from our clustering database make contributions in extracting a library of paratope structural motifs for epitope binding that could be used in prediction or engineering of paratopes on antibody structures. |
|
|
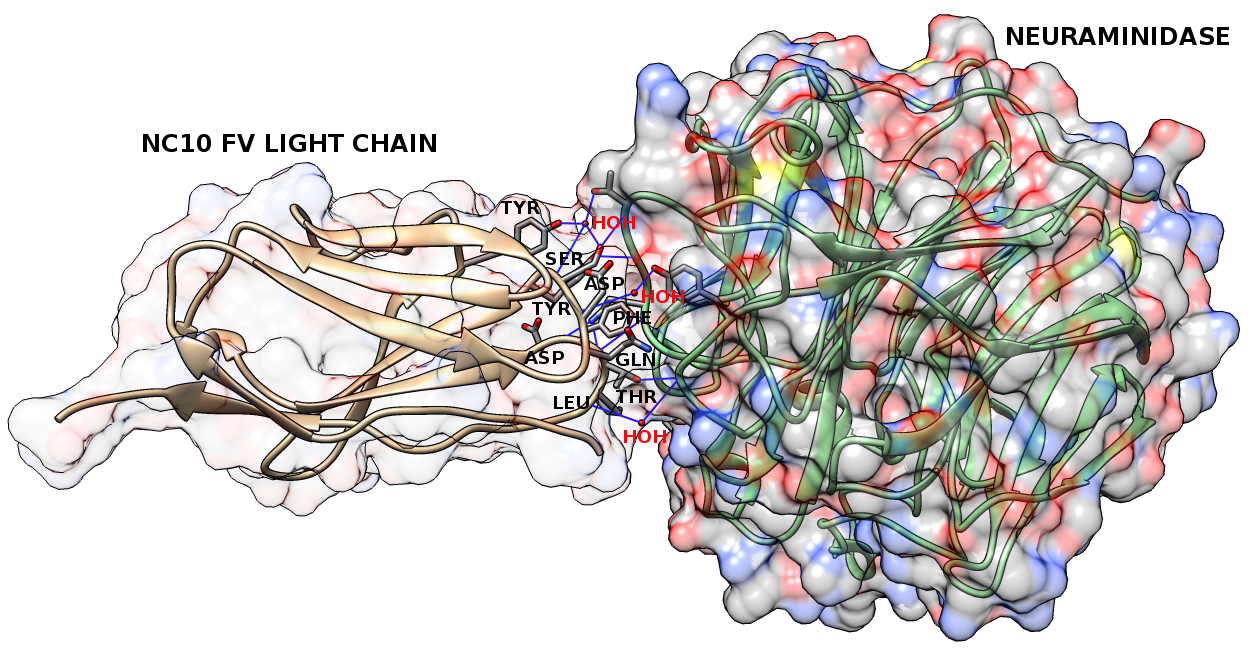
Figure1: The contact residues (gray) on the light chain (chain L; brown) of the anti-neuraminidase antibody NC10 (PDB code: 1NMB). The antigen, neuraminidase from influenza virus (chain N), is in green. The solid regions show the surface of this complex. We identify 9 contact residues from this antibody: Gln 27, Asp 28, Ser 30, Tyr 32, Tyr 50, Asp 91, Phe 92, Thr 93, Leu 94. The blue lines show hydrogen bonds that these contact residues make with antigen residues and/or interfacial water molecules. There are 5 contact residues (Ser 30, Tyr 32, Asp 91, Phe 92, Thr 93) that make 5 hydrogen bonds with antigen residues, 4 contact residues (Asp 28, Ser 30, Tyr 50, Leu 94) that make 5 hydrogen bonds with interfacial water molecules, 8 contact residues (Gln 27, Asp 28, Ser 30, Tyr 32, Asp 91, Phe 92, Thr 93, Leu 94) that participate in van der Waals interactions, 3 contact residues (Tyr 32, Phe 92, Leu 94) involved in hydrophobic interactions and 0 contact residues involved in ionic interactions. |