

Analysis, comparison, visualization of contact residues & interfacial waters of antibody-antigen structures/models
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
Contents
2. Query structures for comparison 3. Output of analyis of contact residues and interfacial waters 4. Output of 3D structure comparison of contact residues 5. Output of browsing antibody-antigen structures from PDB 6. Mouse controls to measure distances, display atom names, and move structures 7. Antibody-antigen complexes 8. Browser compatibility 1. Query structure for analysis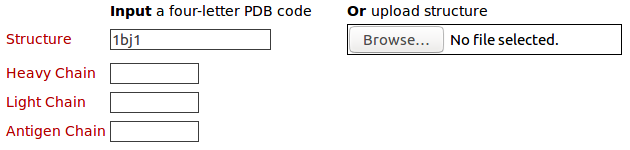
The query structure for antibody drug Avastin (PDB code: 1BJ1) Please click here to look at the output of this query example.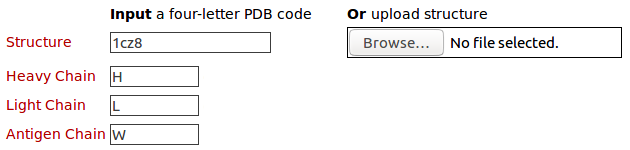
The query structure for antibody drug Lucentis (PDB code: 1CZ8) with heavy chain (H), light chain (L), and antigen chain (W) Please click here to look at the output of this query example.
The query structure for antibody drug Lucentis (PDB code: 1CZ8) with heavy chain (H) and antigen chain (W) Please click here to look at the output of this query example.
The query structure for antibody drug Lucentis (PDB code: 1CZ8) with light chain (L) and antigen chain (W) Please click here to look at the output of this query example.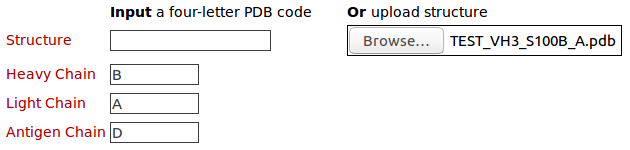
The query structure of 3D model (TEST_VH3_S100B_A.pdb) with heavy chain (B), light chain (A), and antigen chain (D). Click here to download this model. Please click here to look at the output of this query example.2. Query structures for comparison

The query structures of antibody drugs Lucentis (PDB code: 1CZ8) and Avastin (PDB code: 1BJ1) for the 3D structure comparison of their contact residues Please click here to look at the output of this example.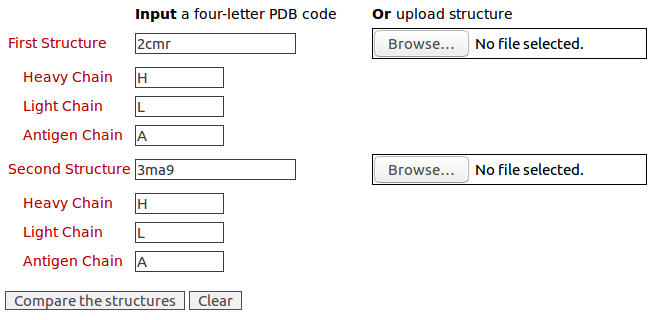
The 3D structure comparison of contact residues of antibodies D5 Fab and Fab 8066 and the antigen HIV-1 gp41 (PDB codes: 2CMR and 3MA9) with heavy chains (H); light chains (L); and antigen chains (A). Please click here to look at the output of this example.
The 3D structure comparison of contact residues of heavy chain of a therapeutic monoclonal antibody (chain H) and its antigen the human interferon alpha-2A (chain D) (PDB code: 4YPG; heavy chain (H); antigen chain (D)) and those of Avastin (PDB code: 1BJ1; heavy chain (H); antigen chain (W)) Please click here to look at the output of this example.3. Output of analyis of contact residues and interfacial waters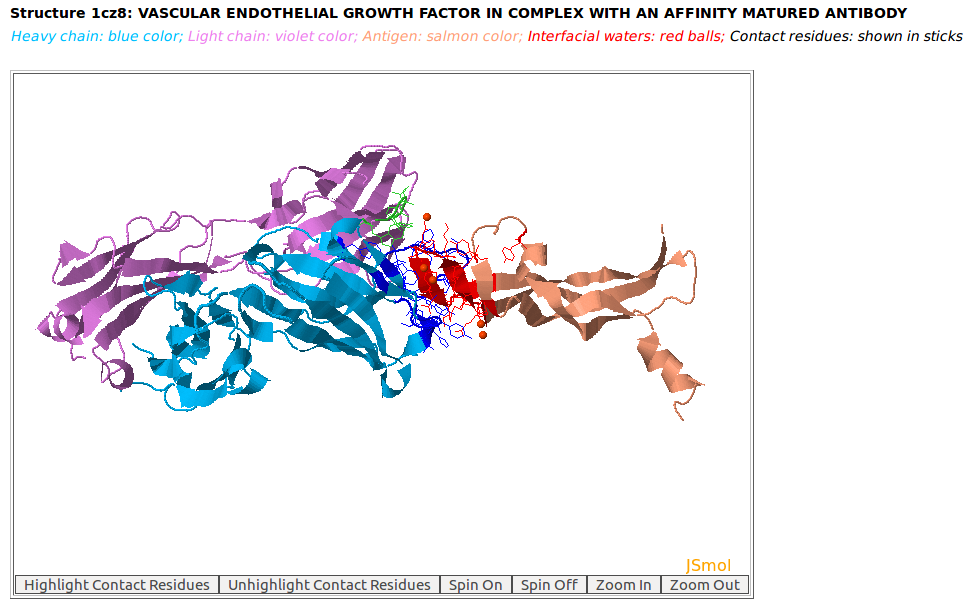
AppA displays the 3D rendition of the antibody drug Lucentis (generic name: ranibizumab, PDB code: 1CZ8; heavy chain: (chain H, blue color); light chain: (chain L, violet color)) with the antigen VEGF (chain W, salmon color) and interfacial waters (red balls) using JSMol (https://jmol.sourceforge.net/). The contact residues of Lucentis and VEGF are highlighted in sticks.
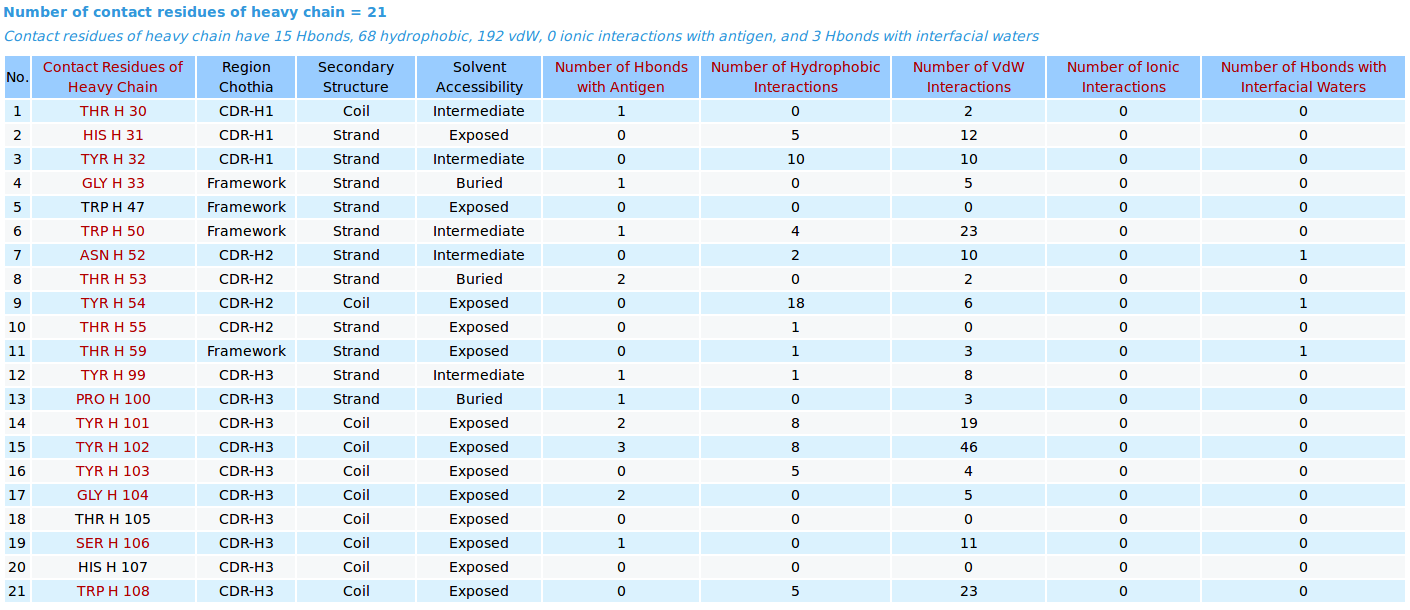

AppA displays the table of contact residues of heavy chain of the antibody drug Lucentis (generic name: ranibizumab, PDB code: 1CZ8) and the antigen VEGF , as well as the number of hydrogen bonds, hydrophobic, van der Waals and ionic interactions that each contact residue is involved in, and their CDR regions, secondary structure and solvent accessibility of contact residues are also shown. In addition to the table, the detailed visualization of contact atoms and interfacial water molecules is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. The detailed hydrogen bonds, hydrophobic, van der Waals and ionic interactions of each contact residue of heavy chain are shown when users click on each contact residue in the table.
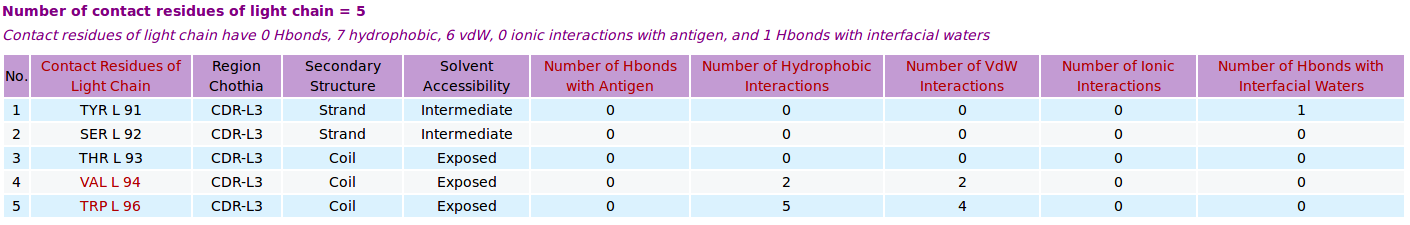

AppA displays the table of contact residues of light chain of the antibody drug Lucentis (generic name: ranibizumab, PDB code: 1CZ8) and the antigen VEGF , as well as the number of hydrogen bonds, hydrophobic, van der Waals and ionic interactions of each contact residue. The detailed visualization of contact atoms and interfacial water molecules is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. The detailed hydrogen bonds, hydrophobic, van der Waals and ionic interactions of each contact residue of light chain are shown when users click on each contact residue in the table.
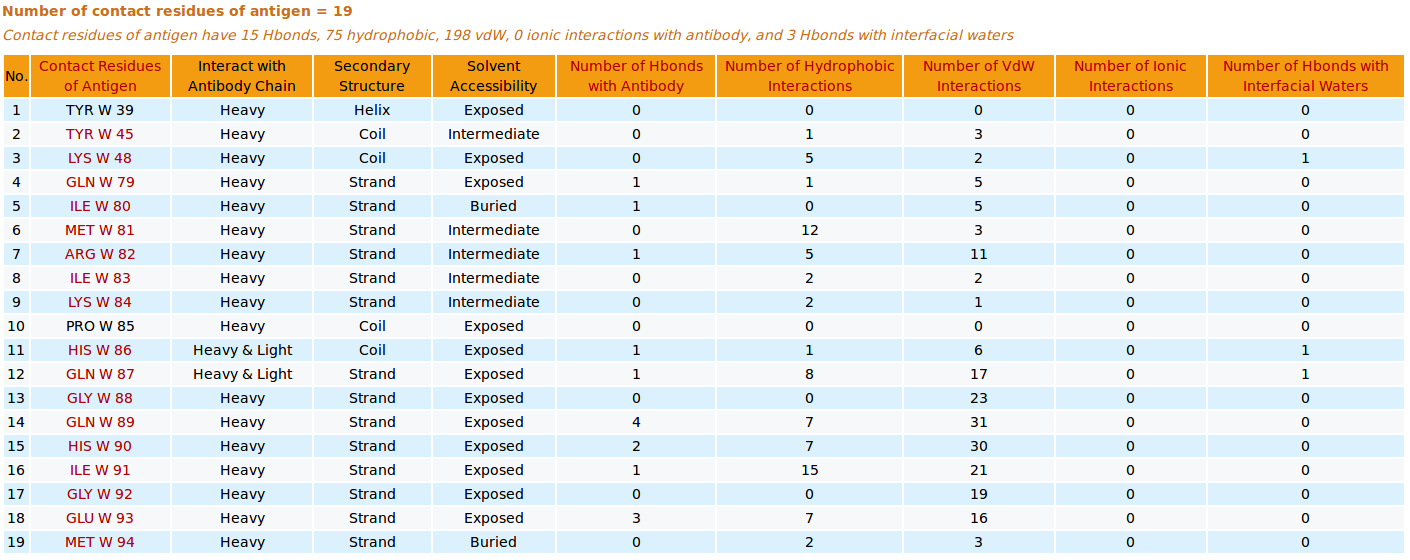
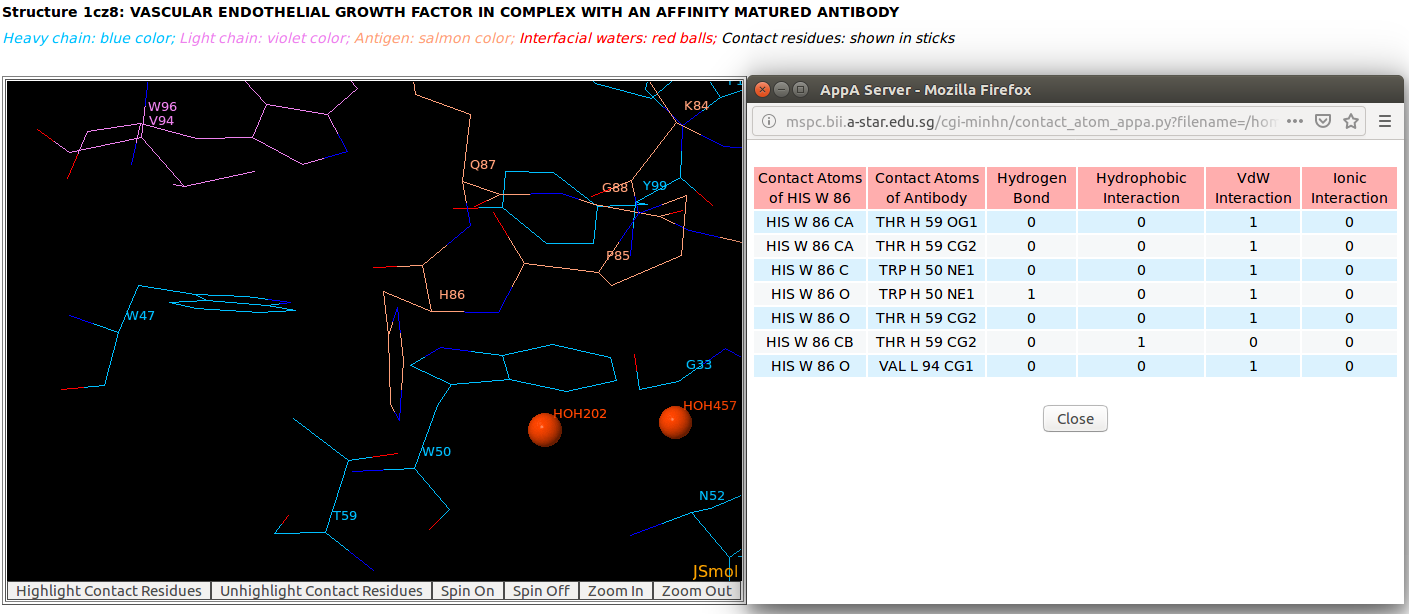
AppA displays the table of contact residues of the antigen VEGF with the antibody drug Lucentis (generic name: ranibizumab, PDB code: 1CZ8). The detailed visualization of contact atoms and interfacial water molecules is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. The detailed hydrogen bonds, hydrophobic, van der Waals and ionic interactions of each contact residue of the antigen VEGF are shown when users click on each contact residue in the table.
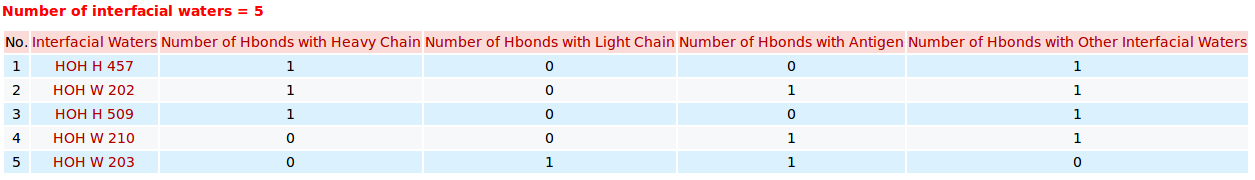
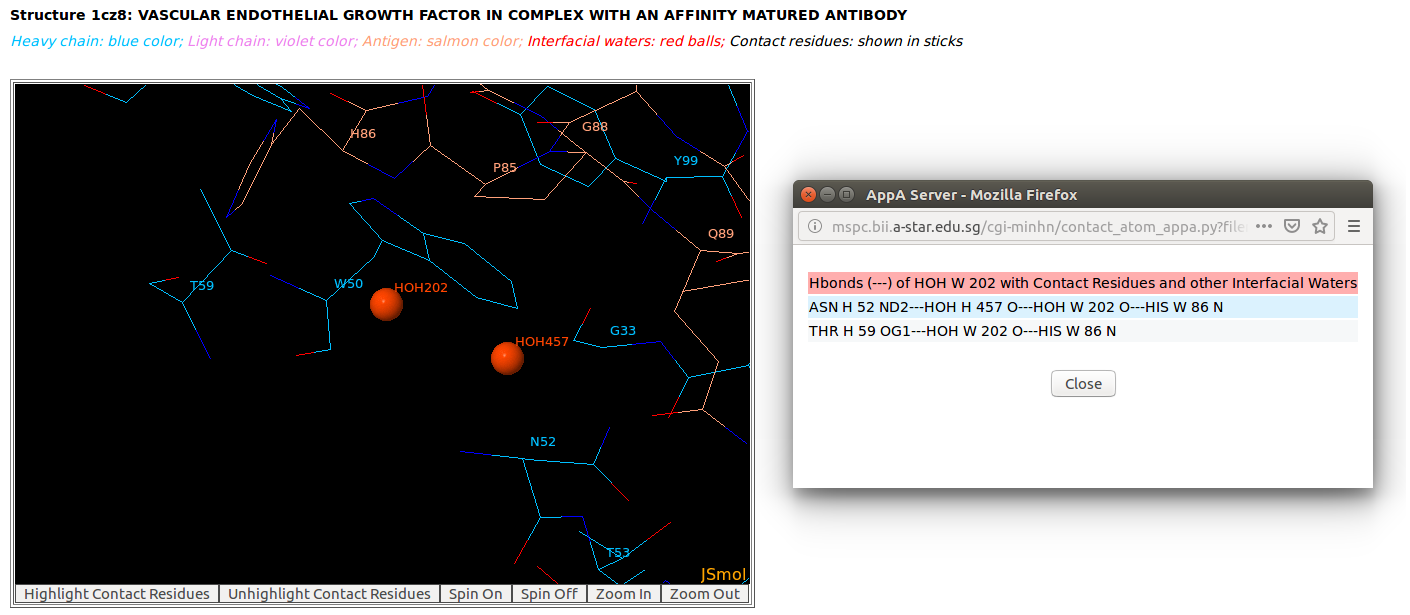
AppA displays the table of interfacial water molecules and their hydrogen bonds with contact residues of antibody drug Lucentis (chains H and L) and antigen VEGF (chain W) of the PDB structure 1CZ8. The detailed visualization of interfacial waters and contact atoms is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. The detailed hydrogen bonds of each interfacial water are shown when users click on each interfacial water in the table.
4. Output of 3D structure comparison of contact residues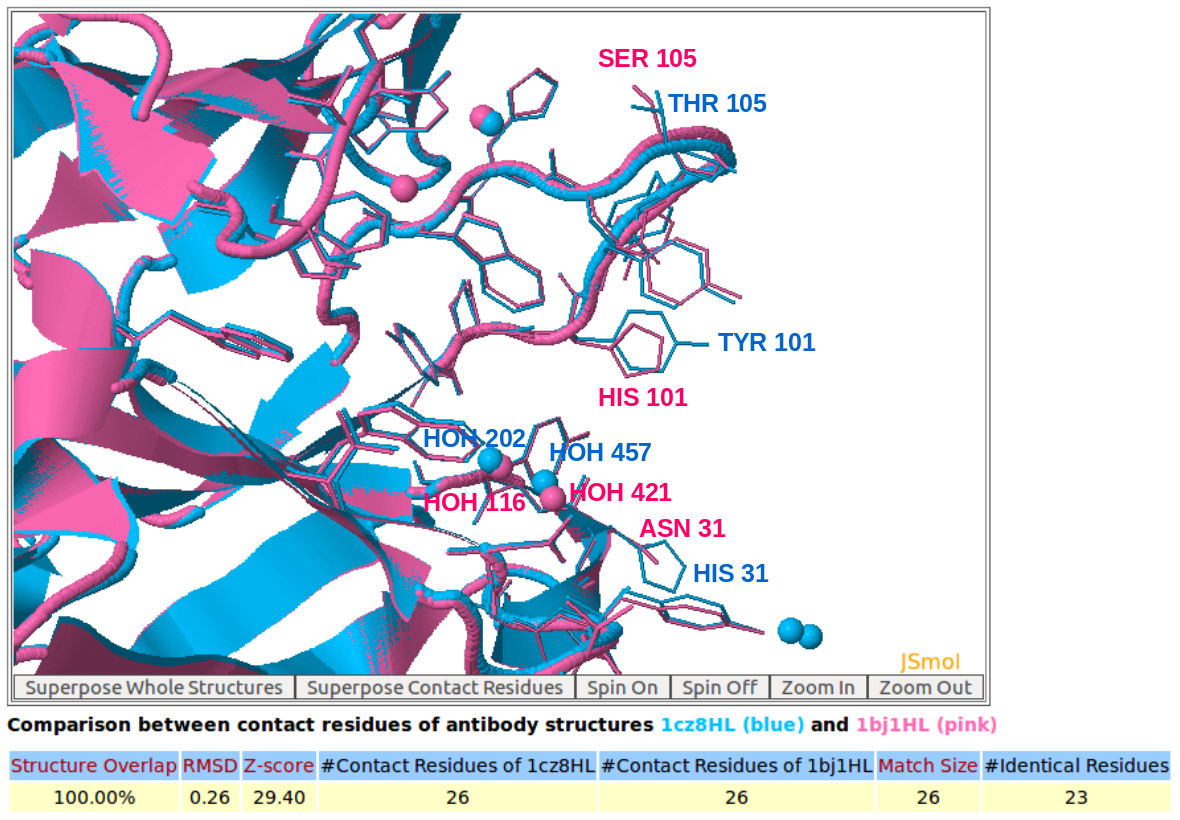
AppA displays the 3D rendition of the structural superimposition of contact residues of heavy chains (H) and light chains (L) of Lucentis (PDB code: 1CZ8) and Avastin (PDB code: 1BJ1) and their interfacial waters using JSMol.
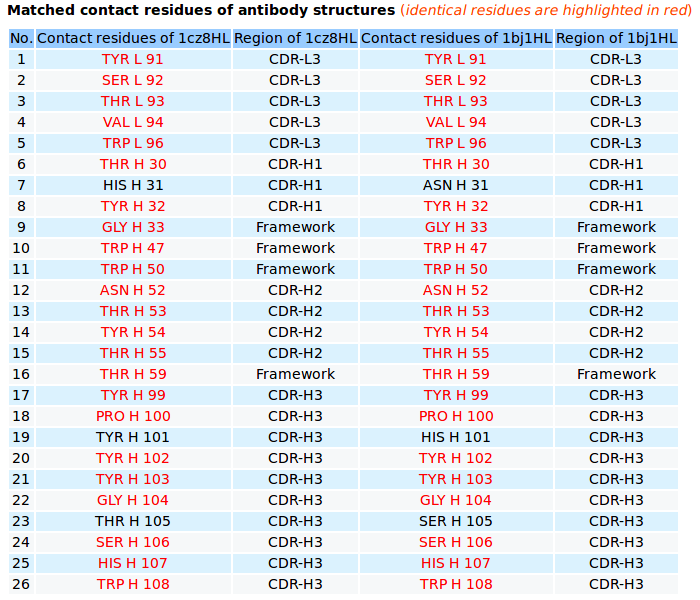
AppA displays the matched contact residues of heavy chains (H) and light chains (L) of Lucentis (PDB code: 1CZ8) and Avastin (PDB code: 1BJ1) for the structure comparison, and the identical matched residues highlighted in red color.
5. Output of browsing antibody-antigen structures from PDB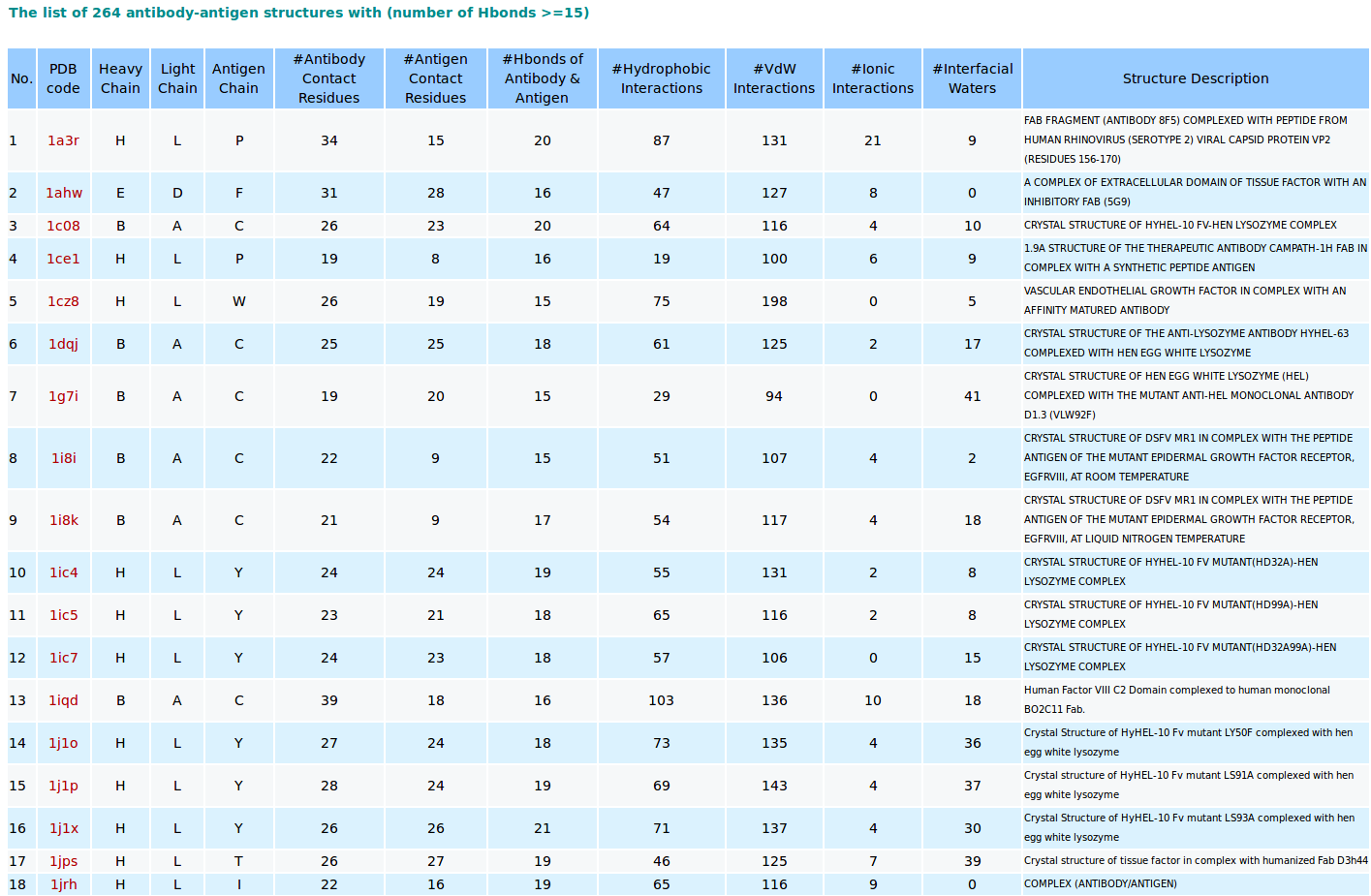
AppA browses existing antibody-antigen structures from the PDB using a cutoff of number of hydrogen bonds of 15. The analysis and visualization of contact residues and interfacial waters of each antibody-antigen complex are shown when users click on its PDB code in the “PDB code” column.
6. Mouse controls for AppA visualization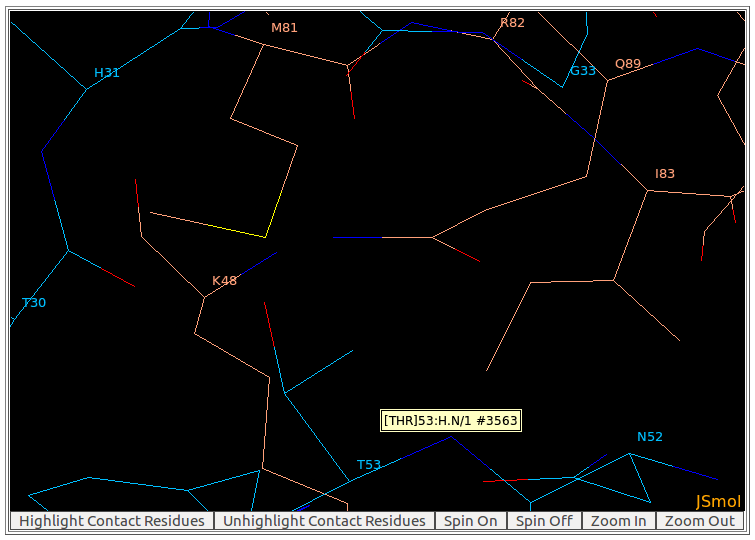
By moving the mouse pointer over any atom in the visualization window of AppA, the corresponding atom name is shown. As seen in the above figure, the name of main chain atom N of THR53 from heavy chain of the antibody drug Lucentis is displayed.
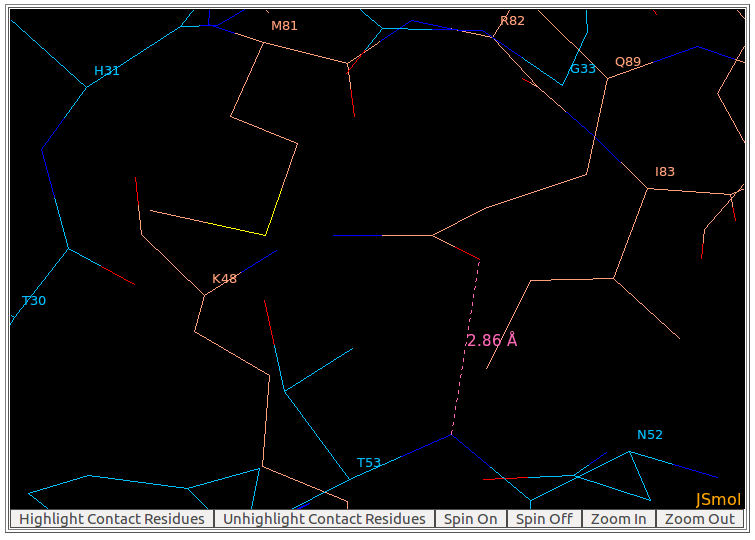
To measure the distance between two atoms, please double click (the left mouse button) on the first atom, and then move the mouse pointer to the second atom. The distance will be shown on a pink line connecting these two atoms. As seen in the above figure, the distance between the main chain atom N of THR53 from heavy chain of the antibody drug Lucentis and the sidechain atom OE1 of GLN89 from antigen VEGF is 2.86Å.
To move the antibody-antigen structure/ contact residues in the visualization window of AppA, please use: CTRL + click and hold the right mouse button. 7. Antibody-antigen complexesThe list of antibody-antigen complexes is updated every week from PDB. Please click here to download the list of PDB codes of antibody-antigen complex structures. 8. Browser compatibility
[back to top] |