

Analysis, comparison, visualization of contact residues & interfacial waters of antibody-antigen structures/models
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
Examples
2. Analysis of contact residues and interfacial waters of 3D structure of antibody drug Lucentis and antigen VEGF 3. Analysis of contact residues and interfacial waters of 3D structure of antibody drug Lucentis and VEGF that is not in SAbDab database 4. Comparison between contact residues of antibody drugs Lucentis and Avastin 5. Comparison between contact residues of a therapeutic monoclonal antibody and antibody drug Avastin 6. Analysis and comparison of contact residues of antibody drugs Pertuzumab (Perjeta) and Trastuzumab (Herceptin) with the antigen HER2 7. Analysis of binding residues of the spike receptor-binding domain of SARS-CoV-2 (SARS-CoV-2-RBD) and SARS-CoV-RBD with human ACE2 |
|
1. Analysis of contact residues of 3D model of antibody drug Avastin and antigen VEGF
|

|
|
AppA displays the rendition of 3D model (TEST_VH3_S100B_A.pdb) of antibody drug Avastin and antigen VEGF with heavy chain (chain B, blue color), light chain (chain A, violet) and the antigen VEGF (chain D, salmon color) using JSMol (https://jmol.sourceforge.net/). The contact residues of antibody and antigen are highlighted in sticks. The detailed input for this example is provided in the help page. This 3D model was built by using MODELLER with Ser at position 100B of the heavy chain of Avastin (PDB code: 1BJ1) mutated to Ala. |
[back to top] |
|
|
|
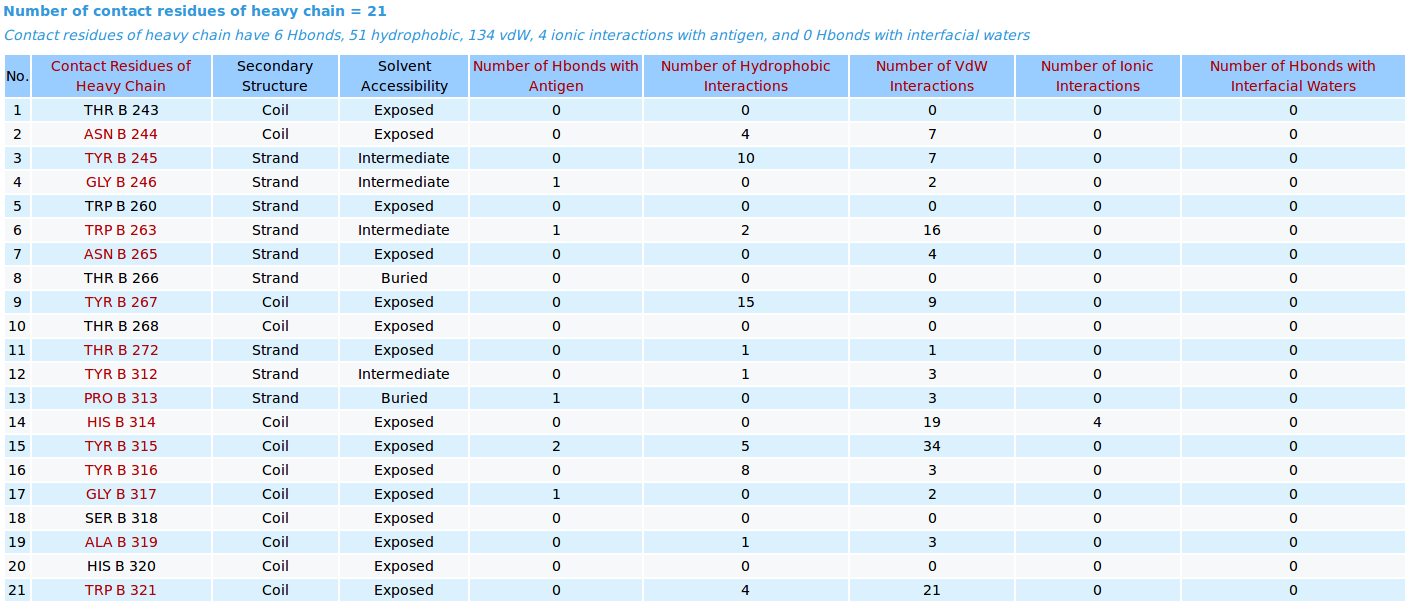
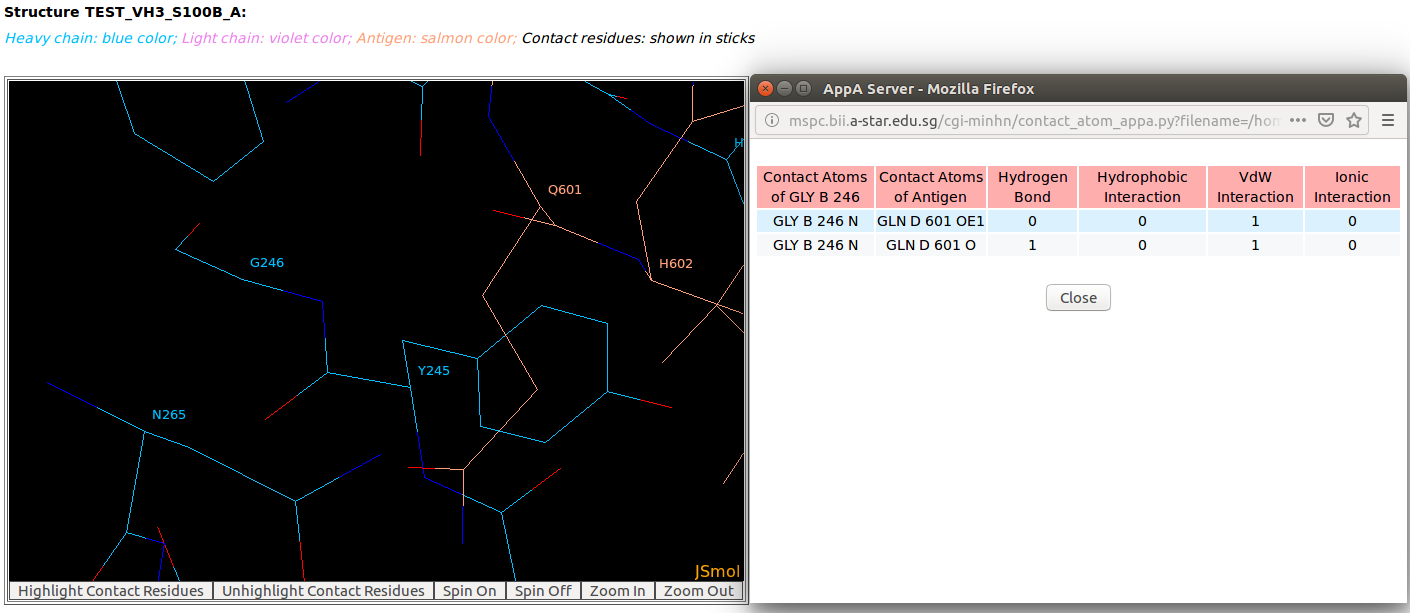
AppA displays the table of contact residues of heavy chain for 3D model (TEST_VH3_S100B_A.pdb) of antibody drug Avastin and antigen VEGF, as well as the number of hydrogen bonds, hydrophobic, van der Waals and ionic interactions that each contact residue is involved in, and their secondary structure and solvent accessibility of contact residues are also shown. In addition to these tables, the detailed visualization of contact atoms is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. The detailed hydrogen bonds, hydrophobic, van der Waals and ionic interactions of each contact residue of heavy chain are shown when users click on each contact residue in the table. As seen, the main chain atom N of GLY246 of heavy chain makes a hydrogen bond with the main chain atom O of GLN601 of the antigen VEGF, and a van der Waals interaction with the sidechain atom OE1 of GLN601 of the antigen. |
[back to top] |
|
|
|
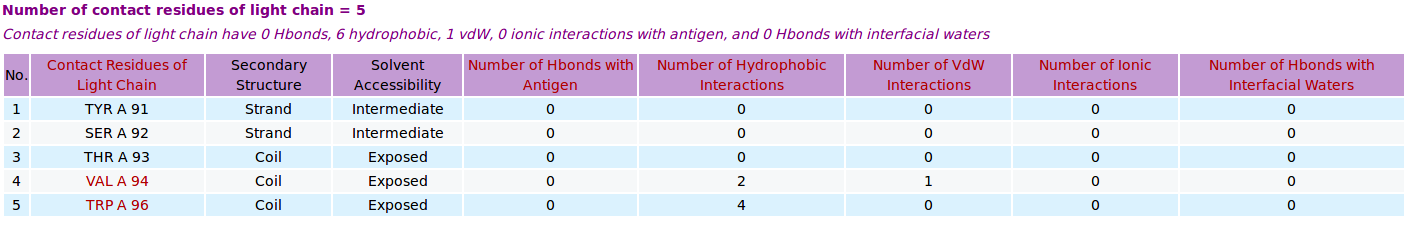
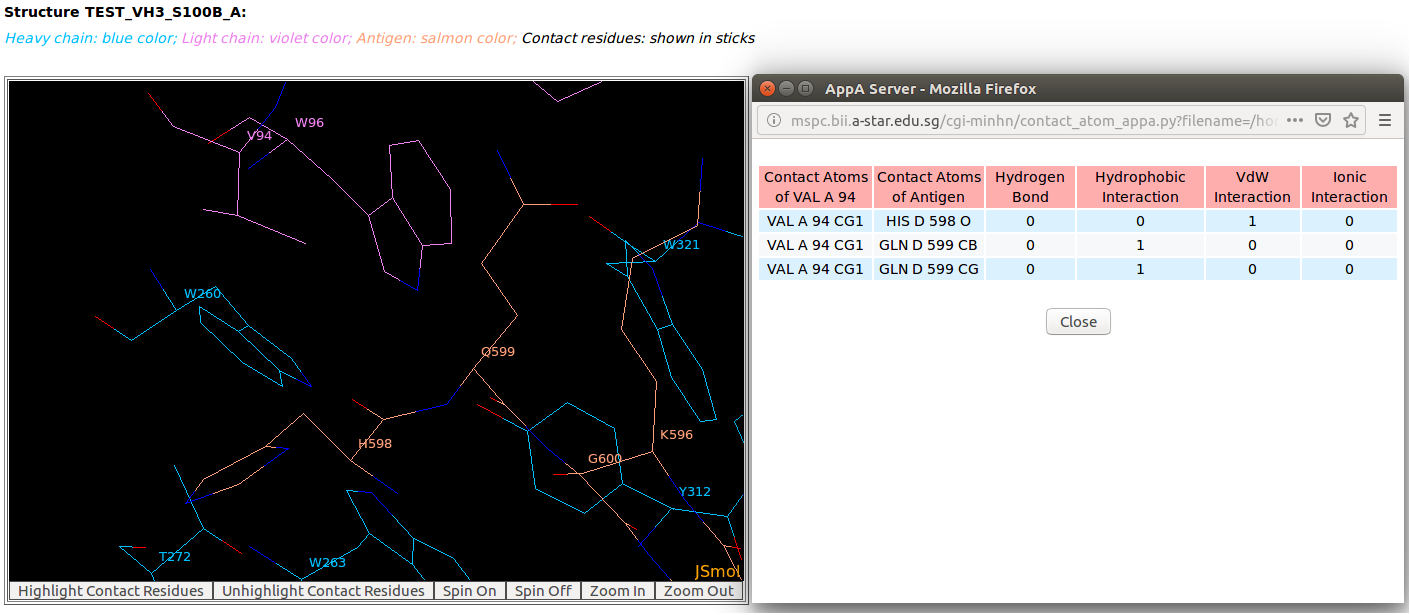
AppA displays the table of contact residues of light chain for 3D model (TEST_VH3_S100B_A.pdb) of antibody drug Avastin and antigen VEGF. The detailed visualization of contact atoms is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. The detailed hydrogen bonds, hydrophobic, van der Waals and ionic interactions of each contact residue of light chain are shown when users click on each contact residue in the table. As seen, the sidechain atom CG1 of VAL94 of light chain makes hydrophobic interactions with the sidechain atoms CB and CG of GLN599 of the antigen VEGF, and a van der Waals interaction with the main chain atom O of HIS598 of the antigen. |
[back to top] |
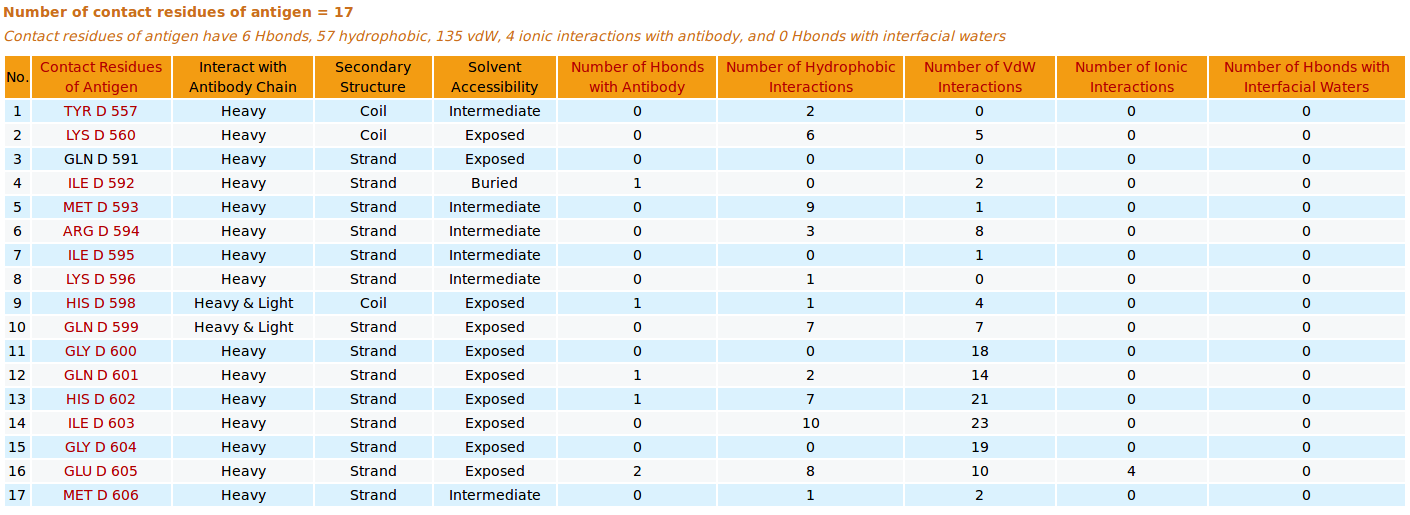
|
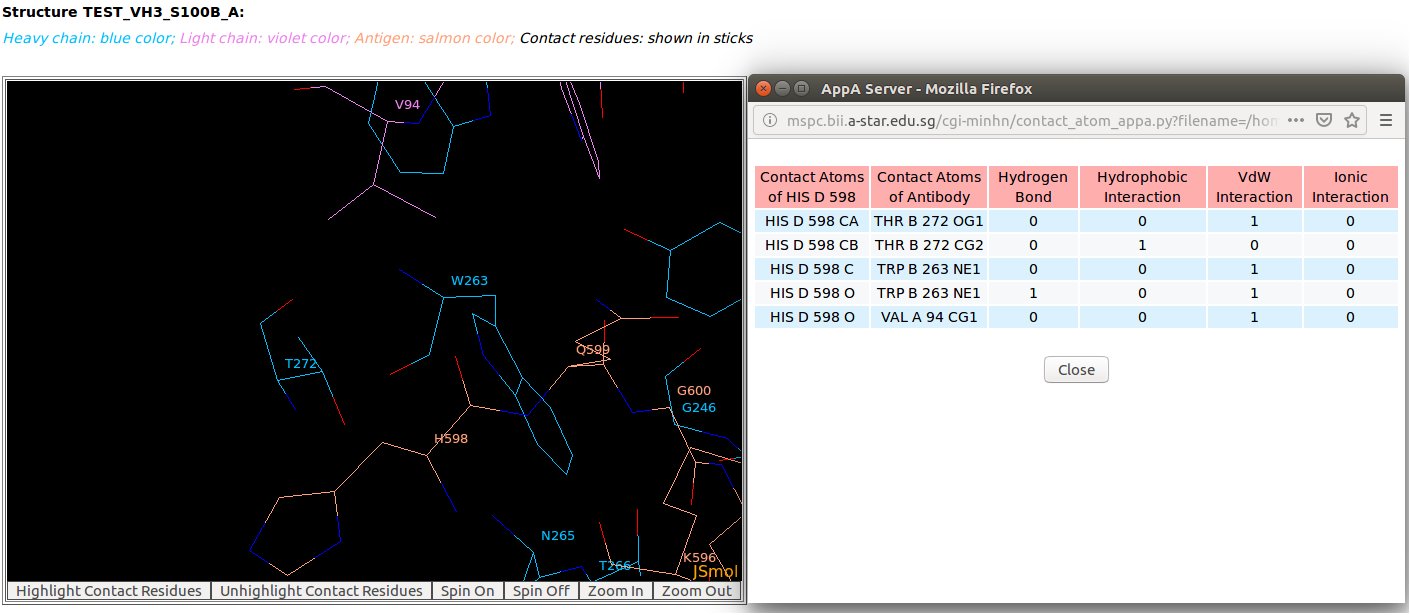
|
|
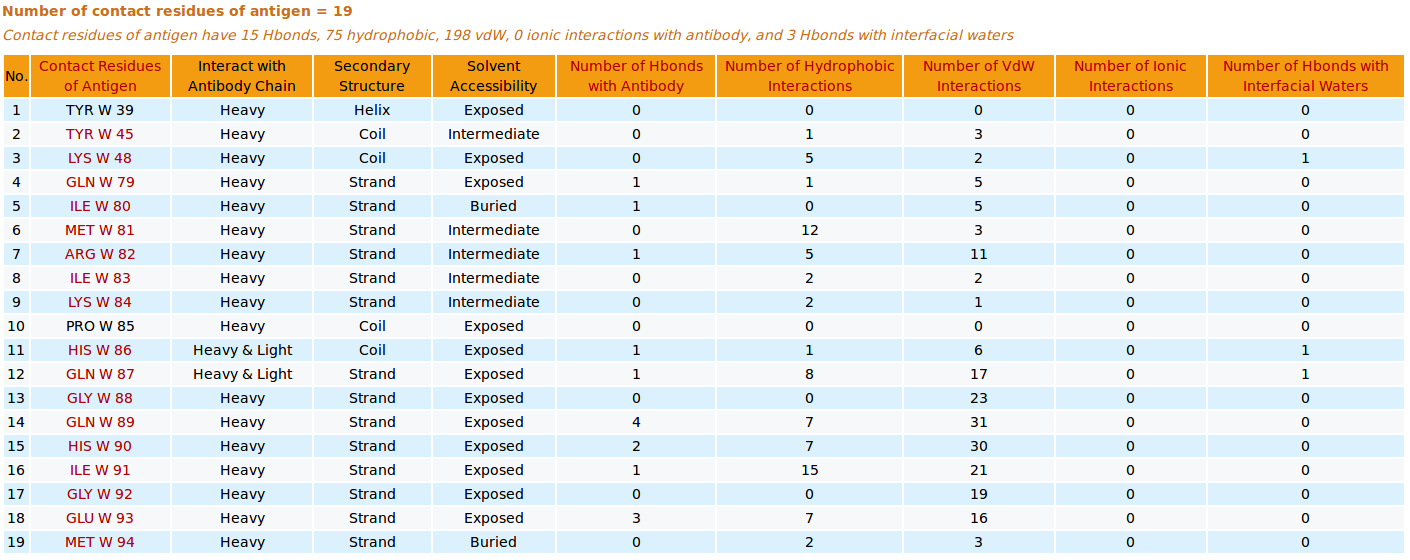
AppA displays the table of contact residues of antigen VEGF for 3D model (TEST_VH3_S100B_A.pdb) with the antibody drug Avastin. The detailed visualization of contact atoms is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. The detailed hydrogen bonds, hydrophobic, van der Waals and ionic interactions of each contact residue of antigen VEGF are shown when users click on each contact residue in the table.
|
[back to top] |
|
2. Analysis of contact residues and interfacial waters of 3D structure of antibody drug Lucentis and antigen VEGF
|
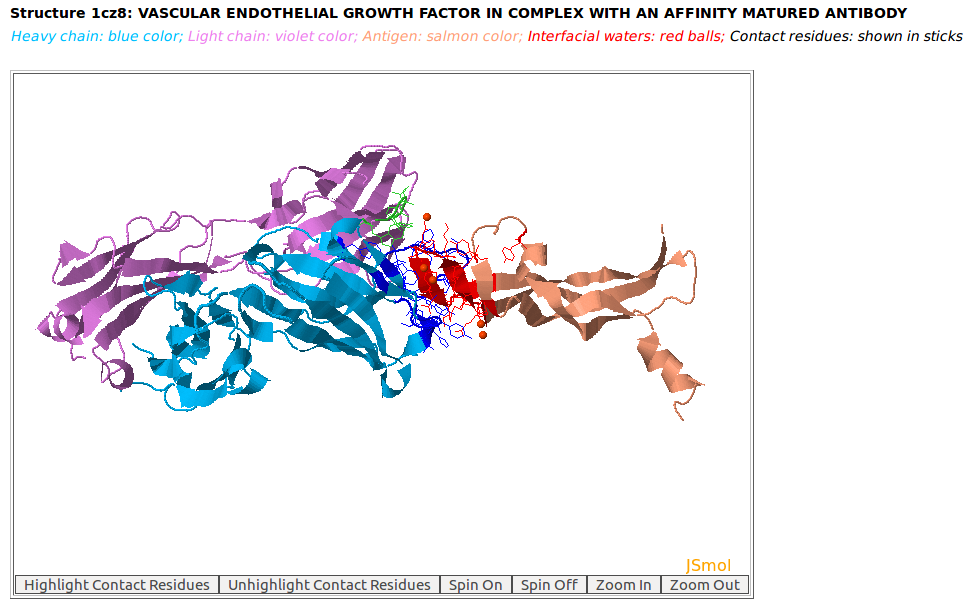
|
|
AppA displays the 3D rendition of the antibody drug Lucentis (generic name: ranibizumab, PDB code: 1CZ8; heavy chain: (chain H, blue color); light chain: (chain L, violet color)) with the antigen VEGF (chain W, salmon color) and interfacial waters (red balls) using JSMol. The contact residues of Lucentis and VEGF are highlighted in sticks. The detailed input for this example is provided in the help page. |
[back to top] |
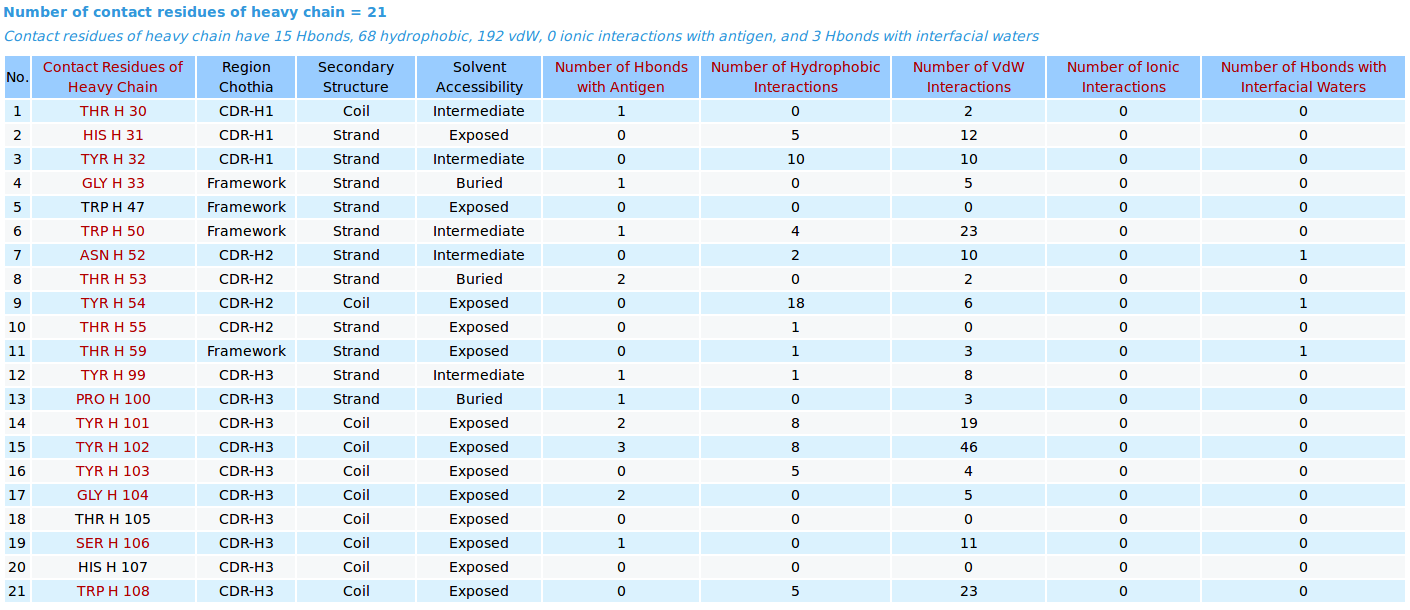
|
|
|
|
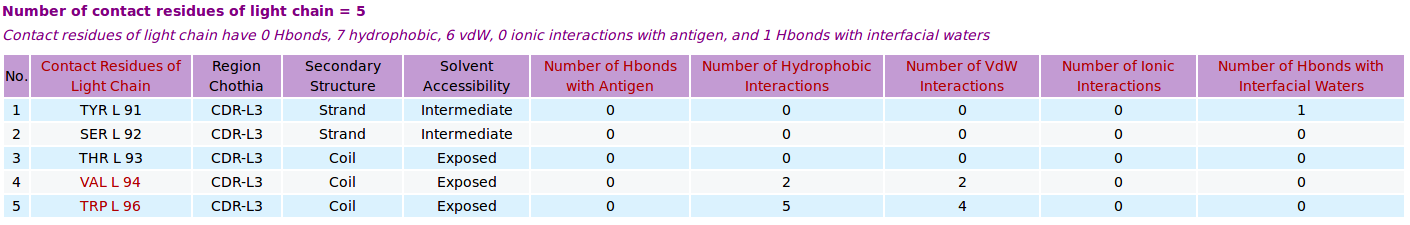
AppA displays the number and list of contact residues of heavy chain (chain H) and light chain (L) of antibody drug Lucentis and antigen VEGF (chain W) of the PDB structure 1CZ8, as well as the number of hydrogen bonds, hydrophobic, van der Waals and ionic interactions and interfacial water molecules that each contact residue is involved in, and their secondary structure and solvent accessibility, and the CDR regions of contact residues of antibody are also shown. |
[back to top] |
|
|
|
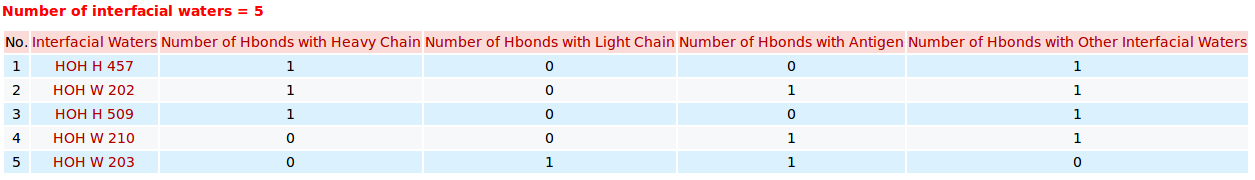
AppA displays the table of interfacial water molecules and their hydrogen bonds with contact residues of antibody drug Lucentis (chains H and L) and antigen VEGF (chain W) of the PDB structure 1CZ8. The detailed visualization of interfacial waters and contact atoms is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. The detailed hydrogen bonds of each interfacial water are shown when users click on each interfacial water in the table. |
[back to top] |
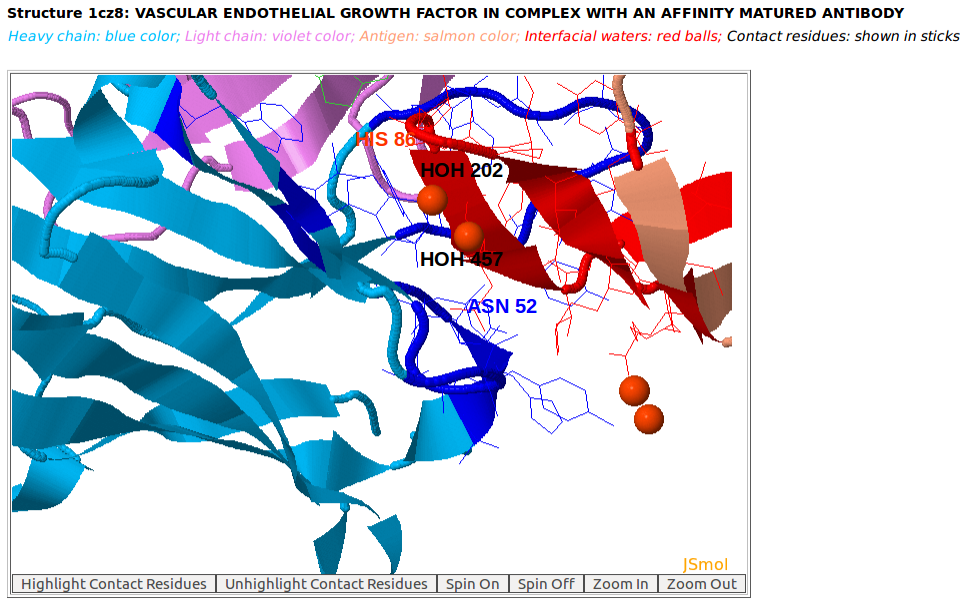
|
|
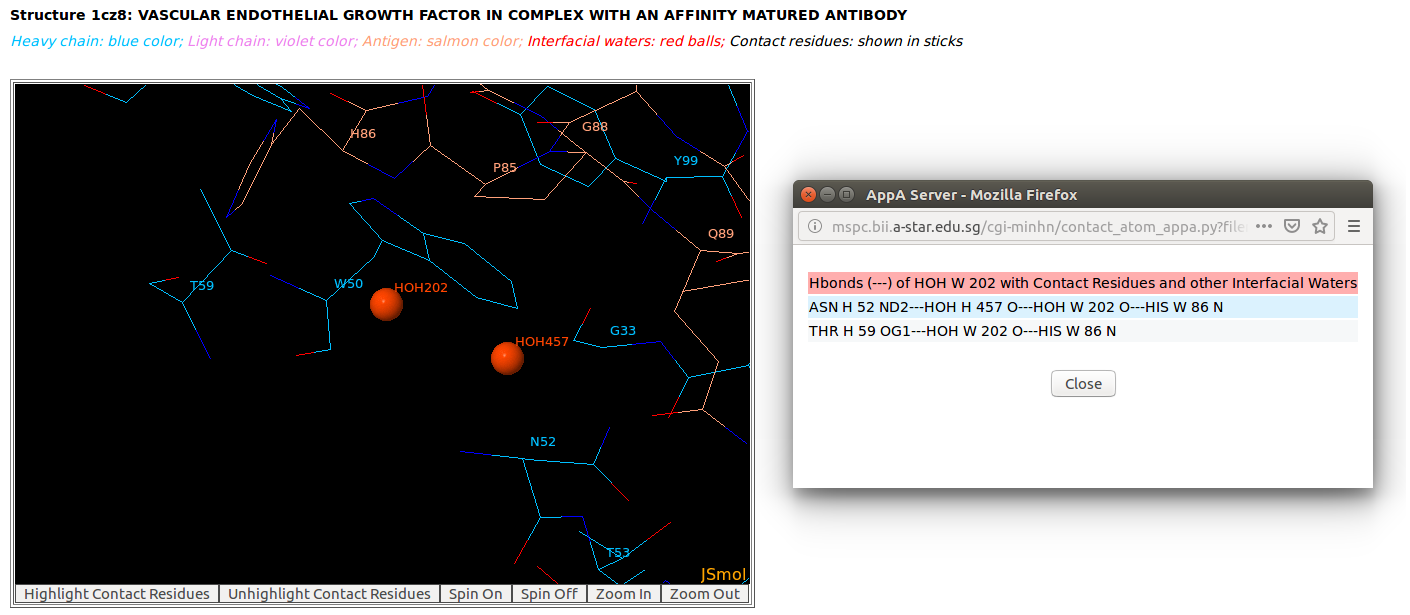
Ordered structures involving interfacial water molecules are observed at the interface of the heavy chain of Lucentis (chain H) and the antigen VEGF (chain W) of the PDB structure 1CZ8 by using "Zoom In" function of AppA.
|
[back to top] |
|
3. Analysis of contact residues and interfacial waters of 3D structure of antibody drug Lucentis and VEGF that is not in SAbDab database
|
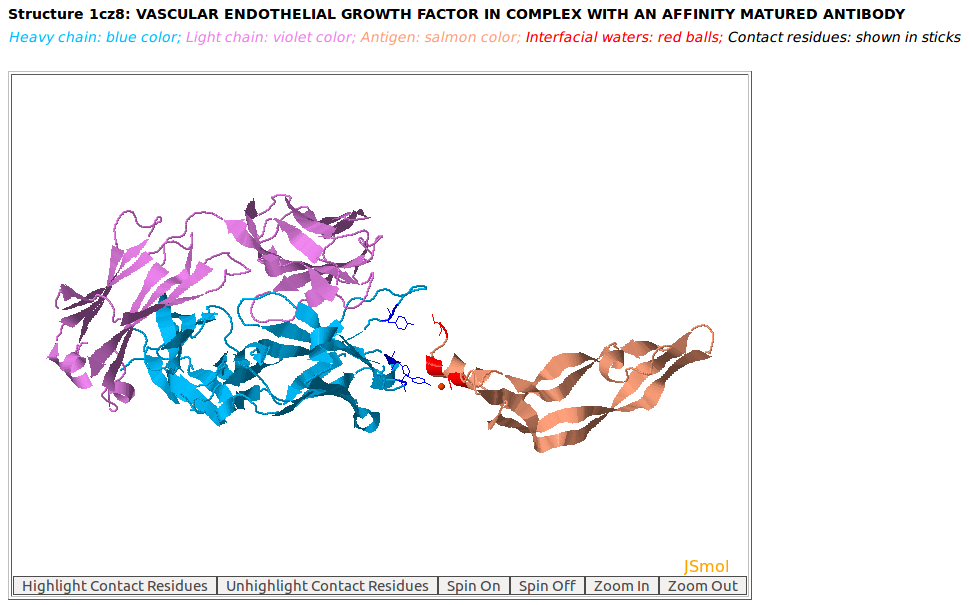
|
|
AppA displays the 3D rendition of the antibody drug Lucentis (PDB code: 1CZ8) with heavy chain (H, blue color), light chain (L, violet) and the antigen VEGF (chain V, salmon color) using JSMol. The input for this example: Structure = 1cz8; Heavy Chain = H; Light Chain = L; Antigen Chain = V; and the complex structure of chains H, L, and V of 1CZ8 is currently not in the list of antibody-antigen structures of SAbDab database. Please note that SAbDab is currently a popular web sever for antibody structures that is available at http://opig.stats.ox.ac.uk/webapps/newsabdab/sabdab/ |
[back to top] |
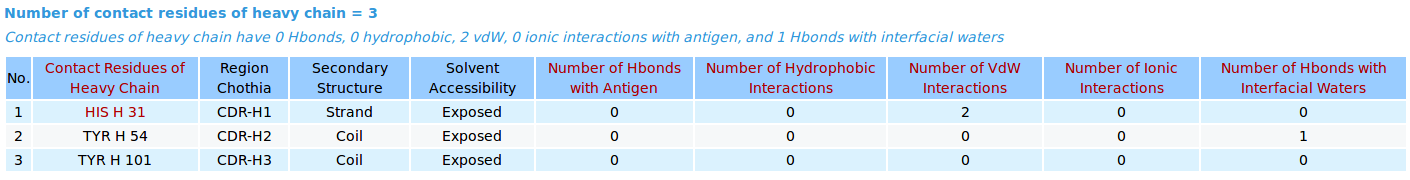
|

|
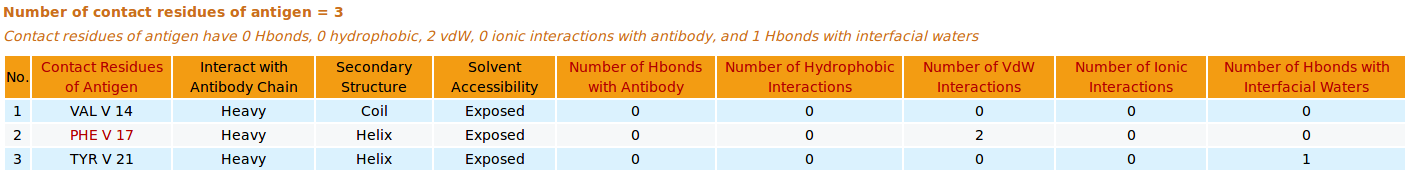
|

|
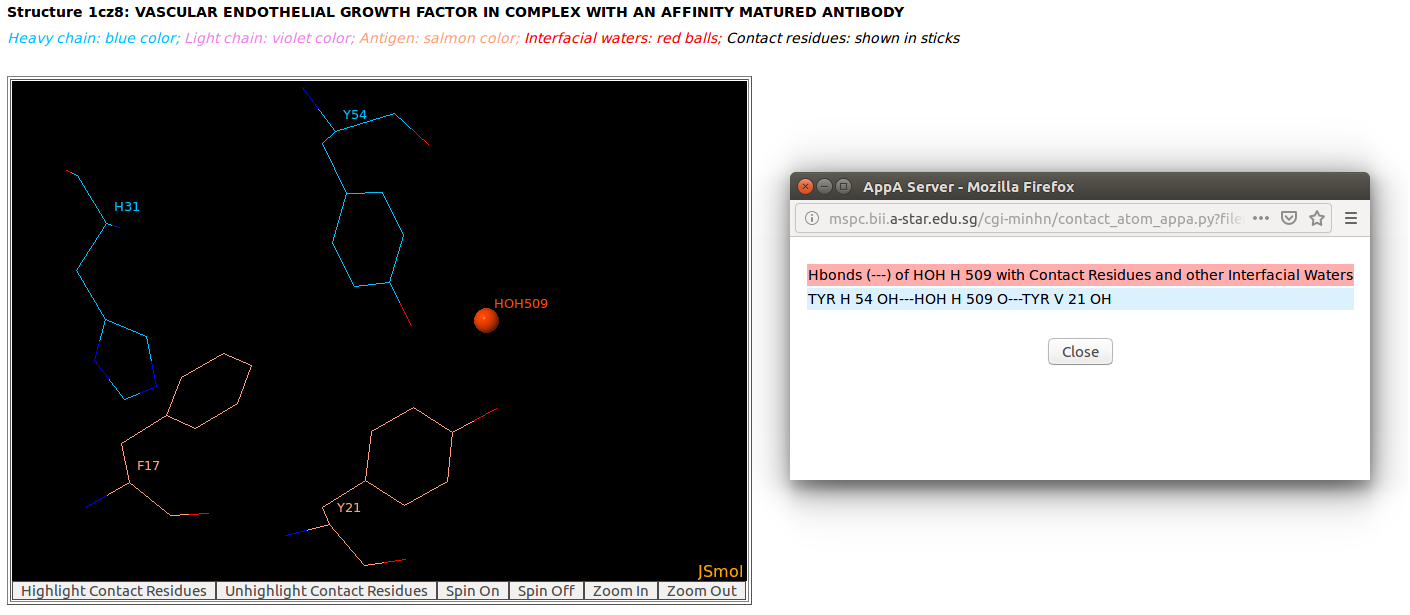
|
|
AppA displays the tables of contact residues of heavy chain (H) and light chain (L) of Lucentis and the antigen VEGF (chain V) of the PDB structure 1CZ8, as well as the number of hydrogen bonds, hydrophobic, van der Waals and ionic interactions and interfacial water molecules that each contact residue is involved in, and their secondary structure and solvent accessibility, and the CDR regions of contact residues of antibody are also shown. The detailed visualization of interfacial waters and contact atoms is displayed using the "Highlight Contact Residues" and "Zoom In" functions of AppA. As seen, the atom O of interfacial water HOH509 make hydrogen bonds with the sidechain atom OH of TYR54 of heavy chain and the sidechain atom OH of TYR21 of antigen VEGF. |
[back to top] |
|
4. Comparison between contact residues of antibody drugs Lucentis and Avastin
|
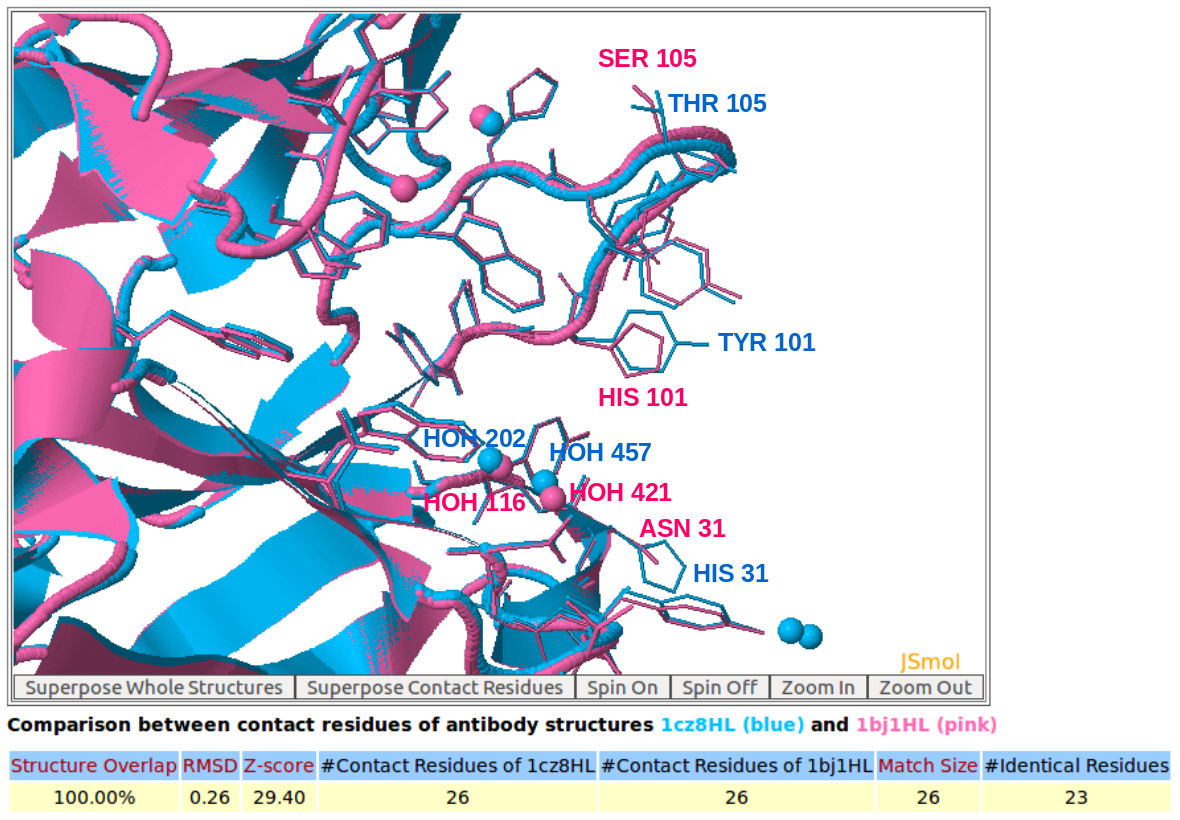
|
|
AppA displays the 3D rendition of the structural superimposition of contact residues of heavy chains (H) and light chains (L) of Lucentis (PDB code: 1CZ8) and Avastin (PDB code: 1BJ1) and their interfacial waters using JSMol. The detailed input for this comparison is provided in the help page.
|
[back to top] |
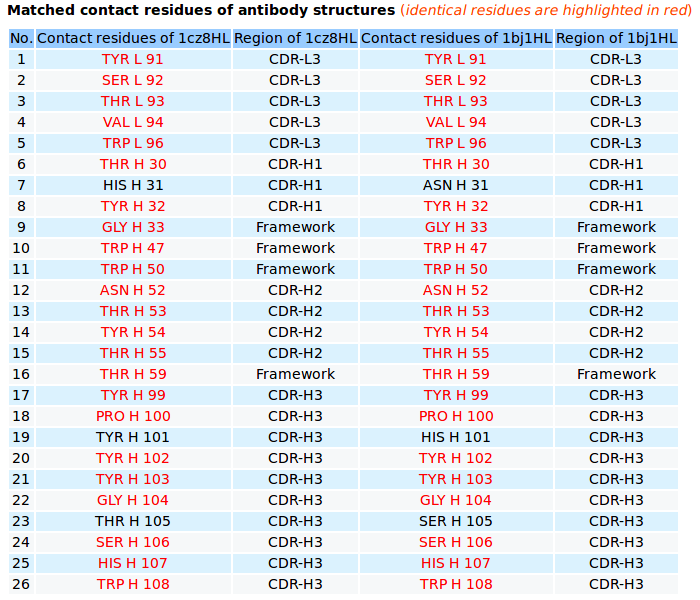
|
|
AppA displays the matched contact residues of heavy chains (H) and light chains (L) of Lucentis (PDB code: 1CZ8) and Avastin (PDB code: 1BJ1) for the structure comparison, and the identical matched residues highlighted in red color. |
[back to top] |
|
5. Comparison between contact residues of a therapeutic monoclonal antibody and antibody drug Avastin
|
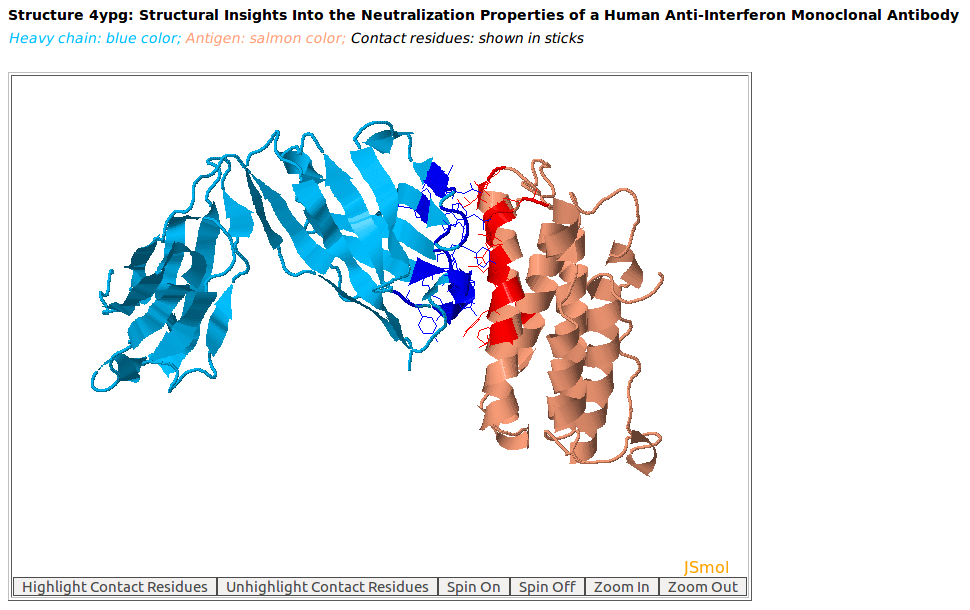
|
|
AppA displays the 3D rendition of the heavy chain of a therapeutic monoclonal antibody (chain H, blue color) with the associated antigen of human interferon alpha-2A (chain D, salmon color) (PDB code: 4YPG) using JSMol. The contact residues of these antibody and antigen are highlighted in sticks. The input for this example: Structure = 4YPG; Heavy Chain = H; Antigen Chain = D |
[back to top] |
|
|
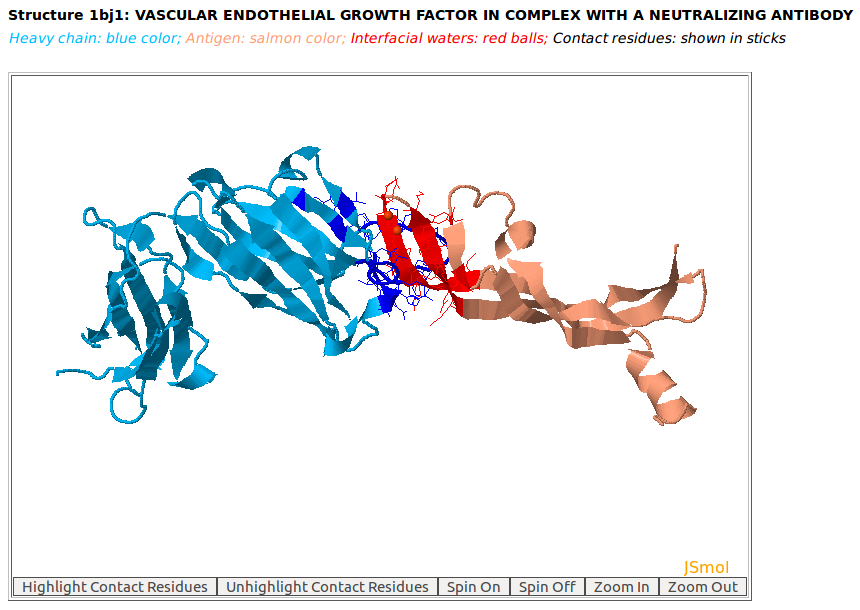
The 3D rendition of the heavy chain of antibody drug Avastin (chain H, blue color) with antigen VEGF (chain W, salmon color) (PDB code: 1BJ1) using JSMol. The contact residues of heavy chain of Avastin and VEGF are highlighted in sticks. The input for this example: Structure = 1BJ1; Heavy Chain = H; Antigen Chain = W |
[back to top] |
|
|
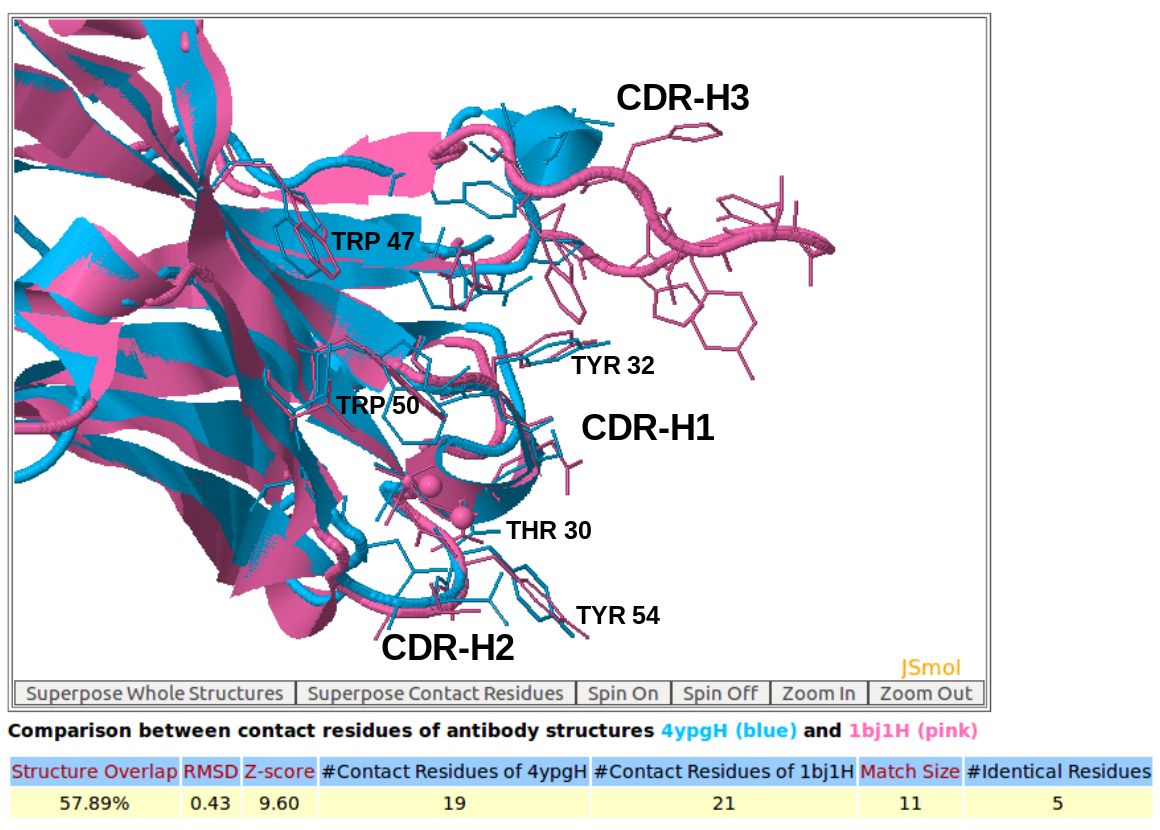
AppA displays the 3D rendition of the structural superimposition of contact residues of the heavy chain (chain H) of the therapeutic monoclonal antibody (PDB code: 4YPG) and the heavy chain (chain H) of Avastin (PDB code: 1BJ1) with their antigens from human interferon alpha-2A (chain D) and VEGF (chain W) using JSMol. The detailed input for this comparison is provided in the help page.
|
[back to top] |
|
6. Analysis and comparison of contact residues of antibody drugs Pertuzumab (Perjeta) and Trastuzumab (Herceptin) with the antigen HER2
|
|
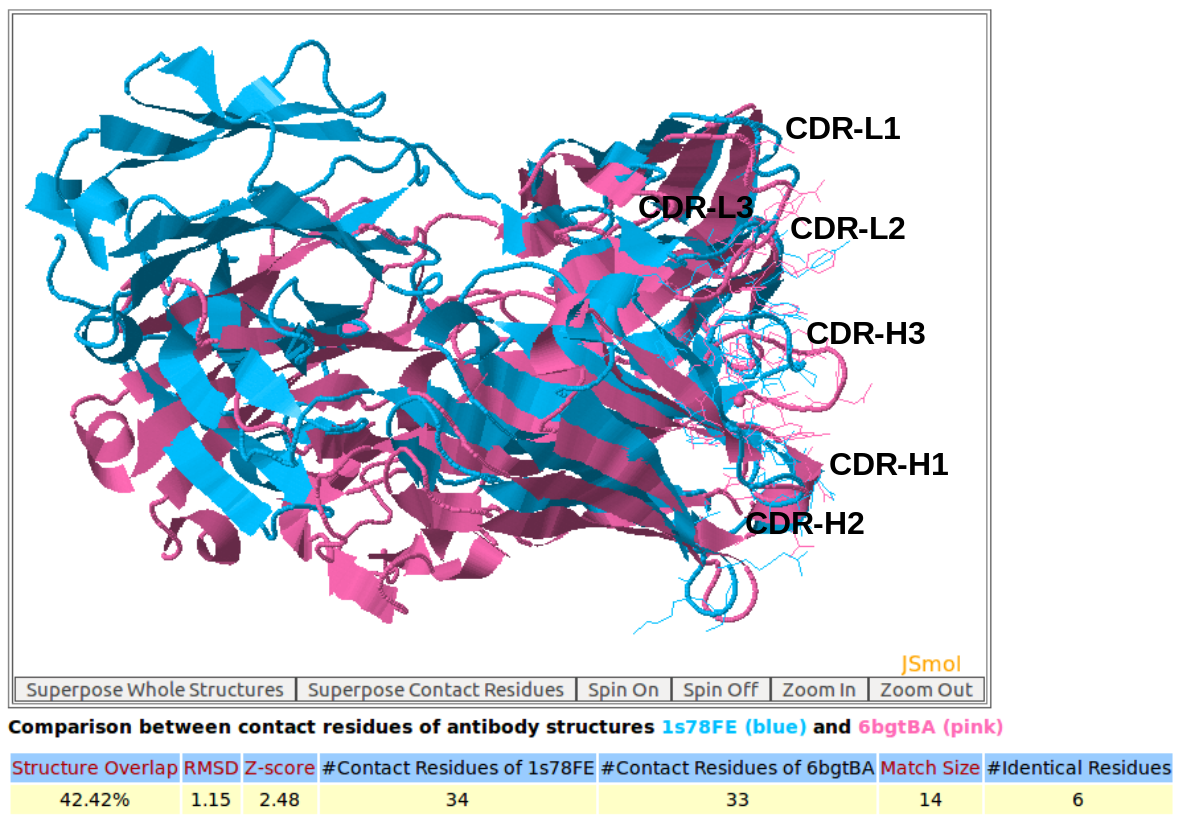
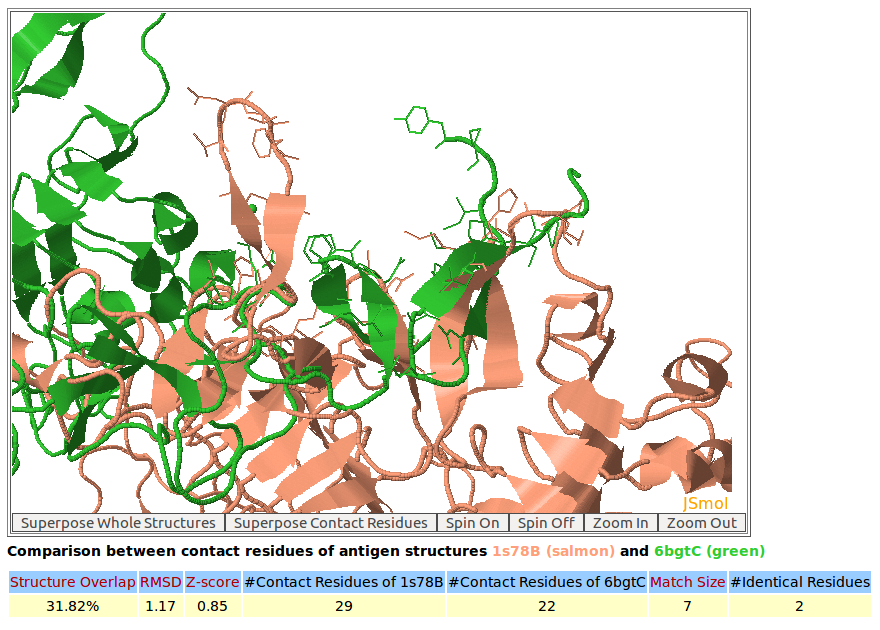
In this example, AppA has been used to analyze and compare the contact residues of antibody drugs Pertuzumab (PDB code: 1S78) and Trastuzumab (PDB code: 6BGT). Both these antibody drugs target the same antigen HER2 (human epidermal growth factor receptor 2) but their binding regions (epitopes) are different (below figures). As seen in the below figures, the comparison of contact residues of antibody drugs Pertuzumab and Trastuzumab shows that their CDR regions (CDR-L1, CDR-L2, CDR-L3, CDR-H1, and CDR-H2) are structurally similar with Z-score of 2.48 while the structures of binding interfaces of their antigens are different with Z-score of 0.85. This study as well as the case study of example 5 indicate that the structures of contact residues from CDR-H1 and CDR-H2 are more conserved compared to those of CDR-H3.
|
|
|
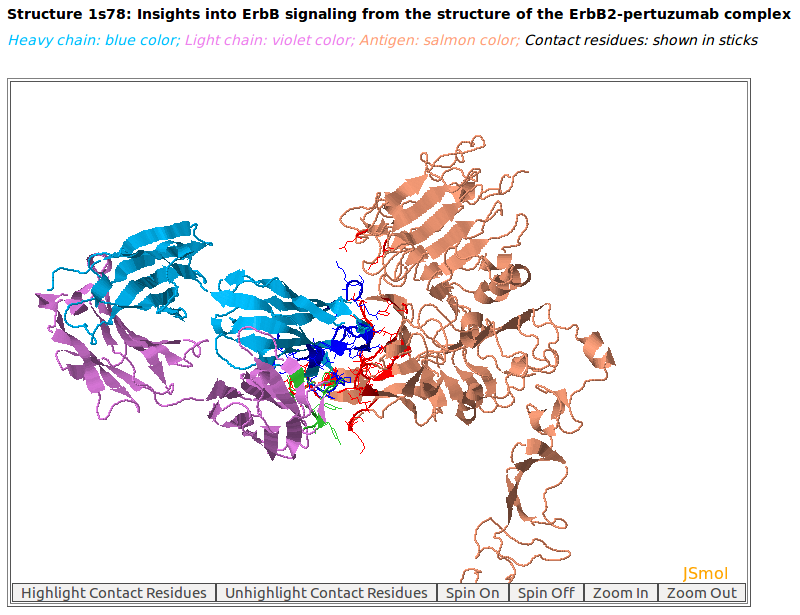
AppA displays the 3D rendition of antibody drug Pertuzumab (blue and violet colors) with the antigen HER2 (salmon color) (PDB code: 1S78). The contact residues of antibody and antigen are highlighted in sticks.
|
|
|
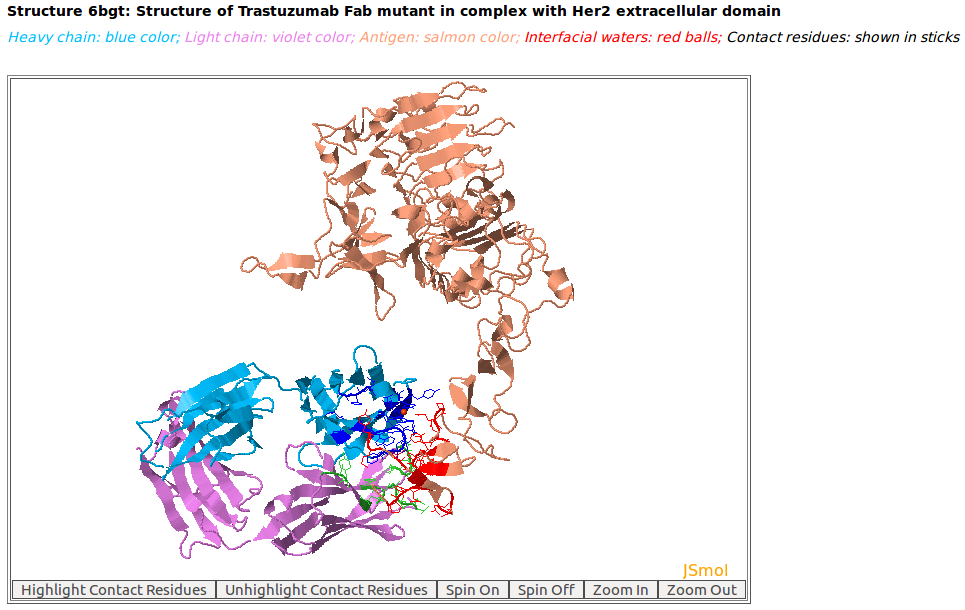
AppA displays the 3D rendition of antibody drug Trastuzumab (blue and violet colors) with the antigen HER2 (salmon color) (PDB code: 6BGT). The contact residues of antibody and antigen are highlighted in sticks. As seen, the binding regions (epitopes) on HER2 of Pertuzumab and Trastuzumab are different.
|
|
|
AppA displays the structural superimposition of contact residues of antibody drugs Pertuzumab (blue color) and Trastuzumab (pink) with their antigens HER2. As seen, the CDR regions (CDR-L1, CDR-L2, CDR-L3, CDR-H1, and CDR-H2) of Pertuzumab and Trastuzumab are structurally similar. The Z-score of the comparison for these antibody contact residues is 2.48. Please click here to look at the output of comparison of contact residues for this example.
|
|
|
AppA displays the structural superimposition of contact residues of antigen HER2 with the antibody drugs Pertuzumab (salmon color) and Trastuzumab (green). The Z-score of the comparison for these antigen contact residues is 0.85. |
[back to top] |
|
7. Analysis of binding residues of the spike receptor-binding domain of SARS-CoV-2 (SARS-CoV-2-RBD) and SARS-CoV-RBD with human ACE2
|
|
AppA has been used not only to analyze and compare the contact residues of antibody-antigen structures, but also can be used for the binding residues of protein-protein complexes. For example, AppA has been used to analyze and compare the binding residues of the spike receptor-binding domain of SARS-CoV-2 (SARS-CoV-2-RBD) and SARS-CoV-RBD with human angiotensin-converting enzyme 2 (ACE2). Please click here to look at the detailed analyses of AppA for binding residues of SARS-CoV-2-RBD and SARS-CoV-RBD with human ACE2. For protein-protein complexes, please download their complexed structures from PDB and then use the option "upload structure/model" of AppA.
|
[back to top] |