

Analysis, comparison, visualization of contact residues & interfacial waters of antibody-antigen structures/models
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
 |
Examples of COVID-19
2. Analysis of binding residues of SARS-CoV-2-RBD of structure 6M0J with human ACE2 3. Analysis of binding residues of SARS-CoV-2-RBD of structure 6LZG with human ACE2 4. Analysis of binding residues of SARS-CoV-RBD of structure 2AJF with human ACE2 5. 3D structure comparison between binding residues of SARS-CoV-2-RBD (6M17) and SARS-CoV-RBD (2AJF) 6. 3D structure comparison between binding residues of SARS-CoV-2-RBD (6M0J) and SARS-CoV-RBD (2AJF) 7. 3D structure comparison between binding residues of SARS-CoV-2-RBD (6LZG) and SARS-CoV-RBD (2AJF) |
| 1. Analysis of binding residues of the spike receptor-binding domain of SARS-CoV-2 (SARS-CoV-2-RBD) of structure 6M17 with ACE2 |
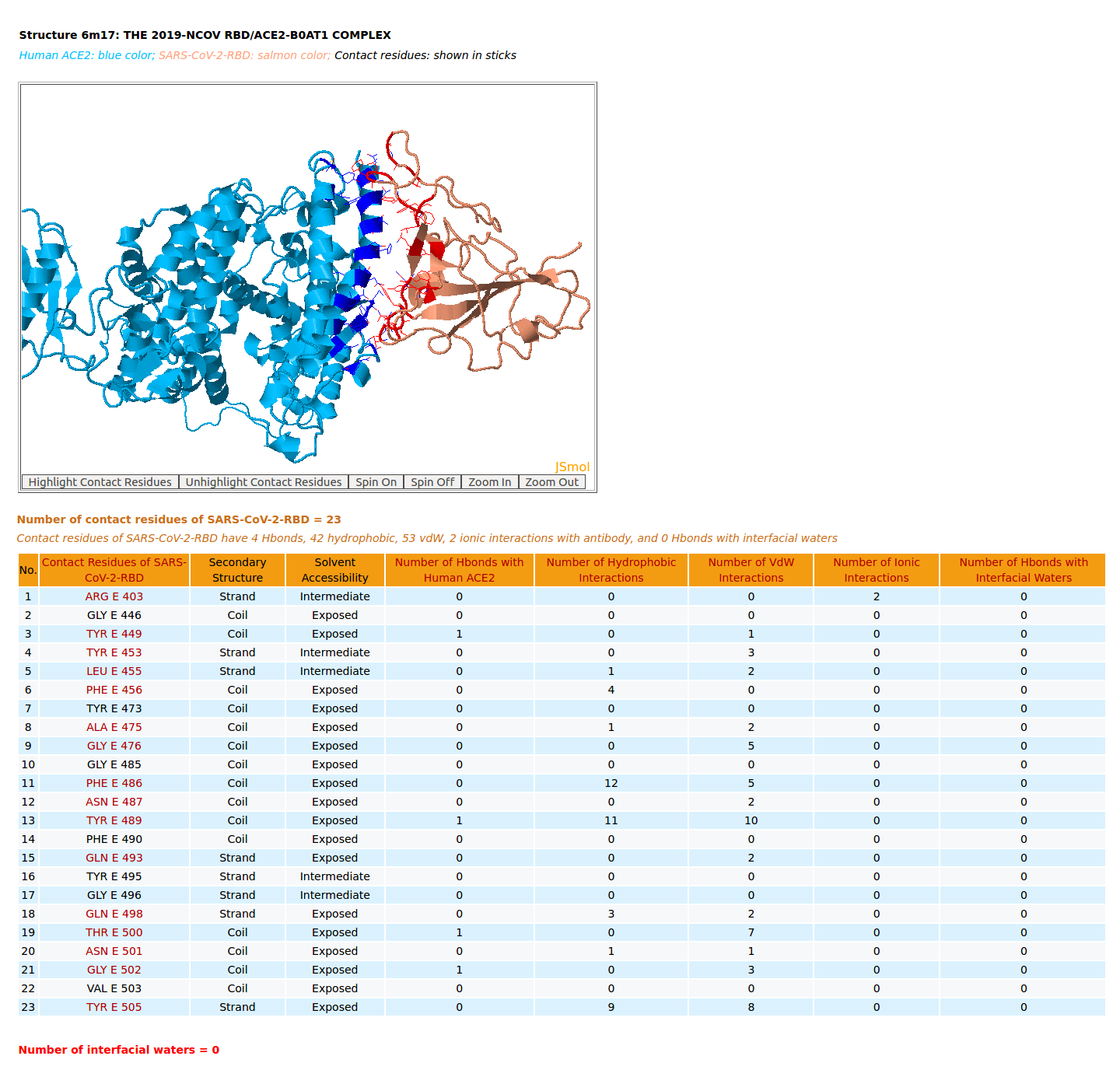
|
|
AppA displays the rendition of 3D structure and the table of of binding residues of the spike receptor-binding domain of SARS-CoV-2 (SARS-CoV-2-RBD) of structure (6M17) with human angiotensin-converting enzyme 2 (ACE2). The binding residues of SARS-CoV-2-RBD are highlighted in salmon sticks. The number of interactions at the interface of SARS-CoV-2-RBD and ACE2 from structure 6M17 (4 Hbonds, 42 hydrophobic, 53 vdW, 2 ionic interactions) are significantly smaller than those of SARS-CoV-RBD and ACE2 (7 Hbonds, 67 hydrophobic, 90 vdW, 4 ionic interactions, 1 Hbond with interfacial waters) from structure 2AJF. The important salt bridge between Lys417 of SARS-CoV-2-RBD and Asp30 of human ACE2 is not found in the structure 6M17 by AppA as the distances between the sidechain atom NZ of Lys417 with OD1 and OD2 of Asp30 are 6.28 Å and 5.67 Å, respectively, which are much larger than the distance criterion of hydrogen bond of 3.5 Å. |
[back to top] |
| 2. Analysis of binding residues of SARS-CoV-2-RBD of structure 6M0J with ACE2 |
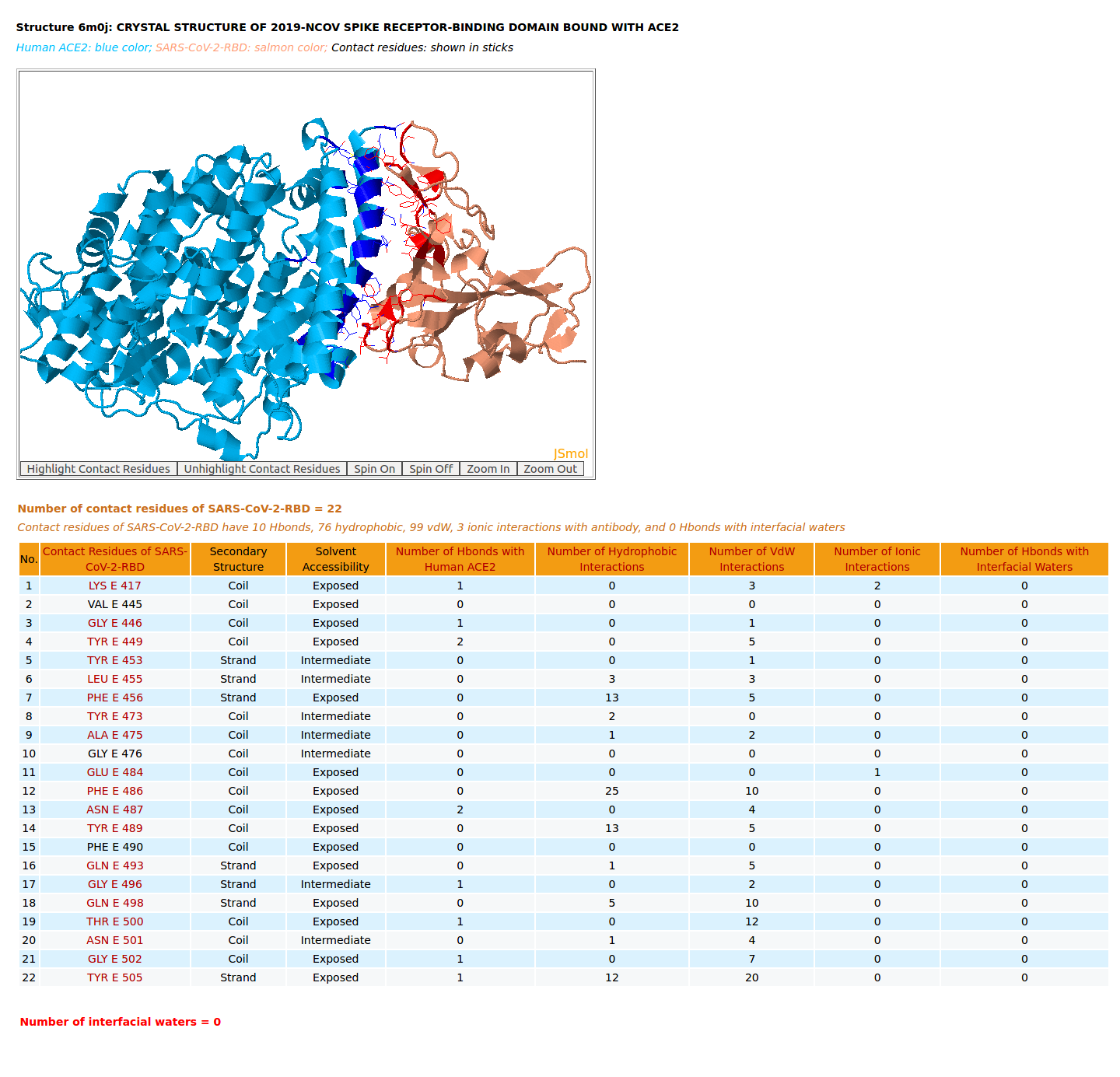
|
|
AppA displays the rendition of 3D structure and the table of of binding residues of SARS-CoV-2-RBD of structure (6M0J) with human ACE2. The binding residues of SARS-CoV-2-RBD are highlighted in salmon sticks. The number of interactions at the interface of SARS-CoV-2-RBD and ACE2 from structure 6M0J (10 Hbonds, 76 hydrophobic, 99 vdW, 3 ionic interactions) are higher than those of SARS-CoV-RBD and ACE2 (7 Hbonds, 67 hydrophobic, 90 vdW, 4 ionic interactions, 1 Hbond with interfacial waters) from structure 2AJF. As shown in the table of binding residues of SARS-CoV-2-RBD of structure 6M0J, the important salt bridge between Lys417 of SARS-CoV-2-RBD and Asp30 of human ACE2 is found in the structure 6M0J by AppA. |
[back to top] |
| 3. Analysis of binding residues of SARS-CoV-2-RBD of structure 6LZG with ACE2 |
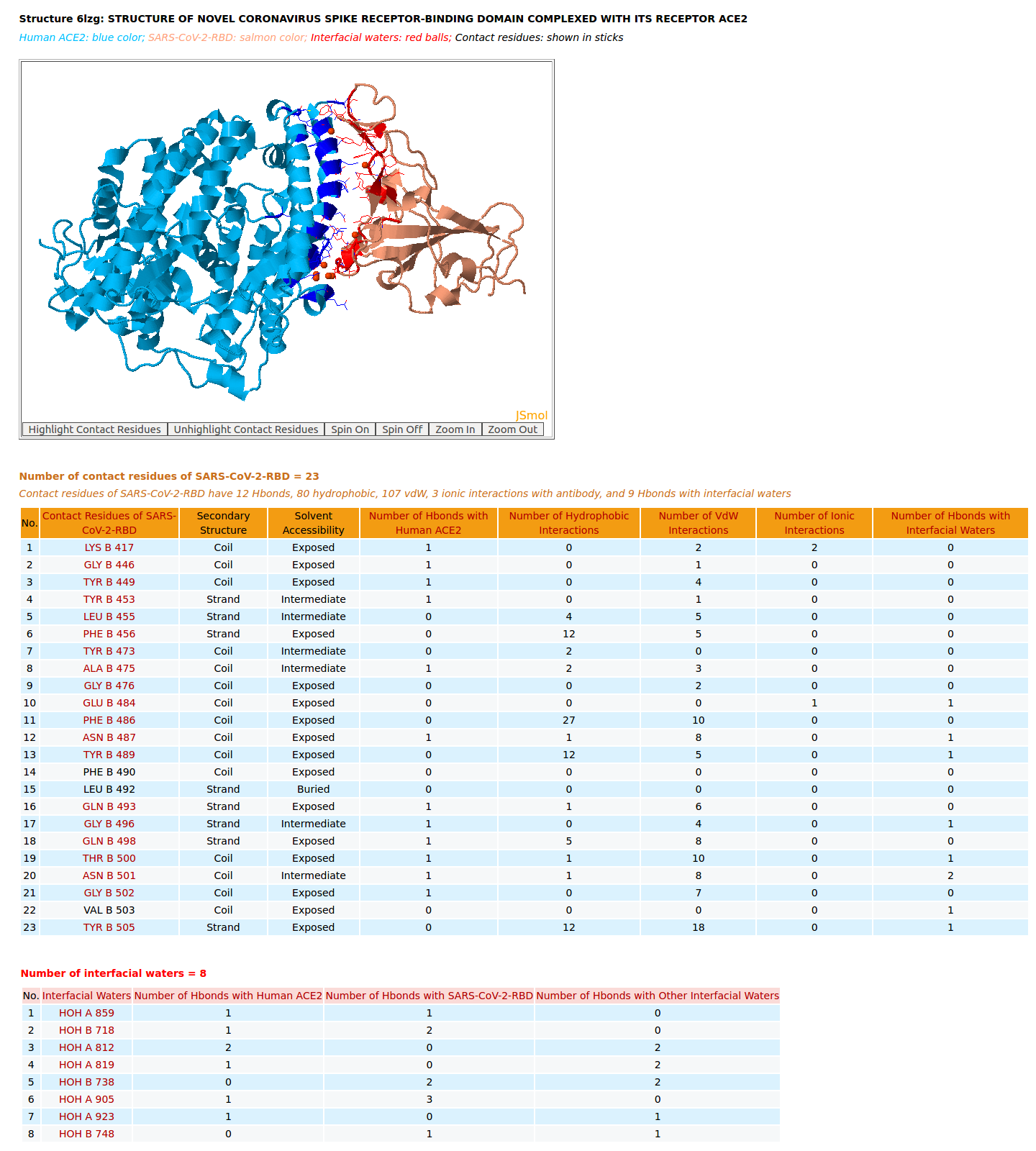
|
|
AppA displays the rendition of 3D structure and the table of of binding residues of SARS-CoV-2-RBD of structure (6LZG) with human ACE2. The binding residues of SARS-CoV-2-RBD are highlighted in salmon sticks. The number of interactions at the interface of SARS-CoV-2-RBD and ACE2 from structure 6LZG (12 Hbonds, 80 hydrophobic, 107 vdW, 3 ionic interactions, 9 Hbonds with interfacial waters) are much higher than those of SARS-CoV-RBD and ACE2 (7 Hbonds, 67 hydrophobic, 90 vdW, 4 ionic interactions, 1 Hbond with interfacial waters) from structure 2AJF. As shown in the table of binding residues of SARS-CoV-2-RBD of structure 6LZG, the important salt bridge between Lys417 of SARS-CoV-2-RBD and Asp30 of human ACE2 is also found in the structure 6LZG by AppA. In addition, among three structures 6M17, 6M0J, and 6LZG only the structure 6LZG could show Hbonds of the critical residues Gln493 and Asn501 of SARS-CoV-2-RBD with Glu35 and Tyr41 of human ACE2. Among three structures 6M17, 6M0J, and 6LZG the analyses from 6M17 do not agree with the binding experiments that have demonstrated that the affinity SARS-CoV-2-RBD for ACE2 is higher than that of SARS-CoV-RBD for ACE2. Meanwhile, the number of interactions at the interface of SARS-CoV-2-RBD and ACE2 from the structure 6M0J and 6LZG agree well with the binding experiments. |
[back to top] |
| 4. Analysis of binding residues of SARS-CoV-RBD of structure 2AJF with ACE2 |
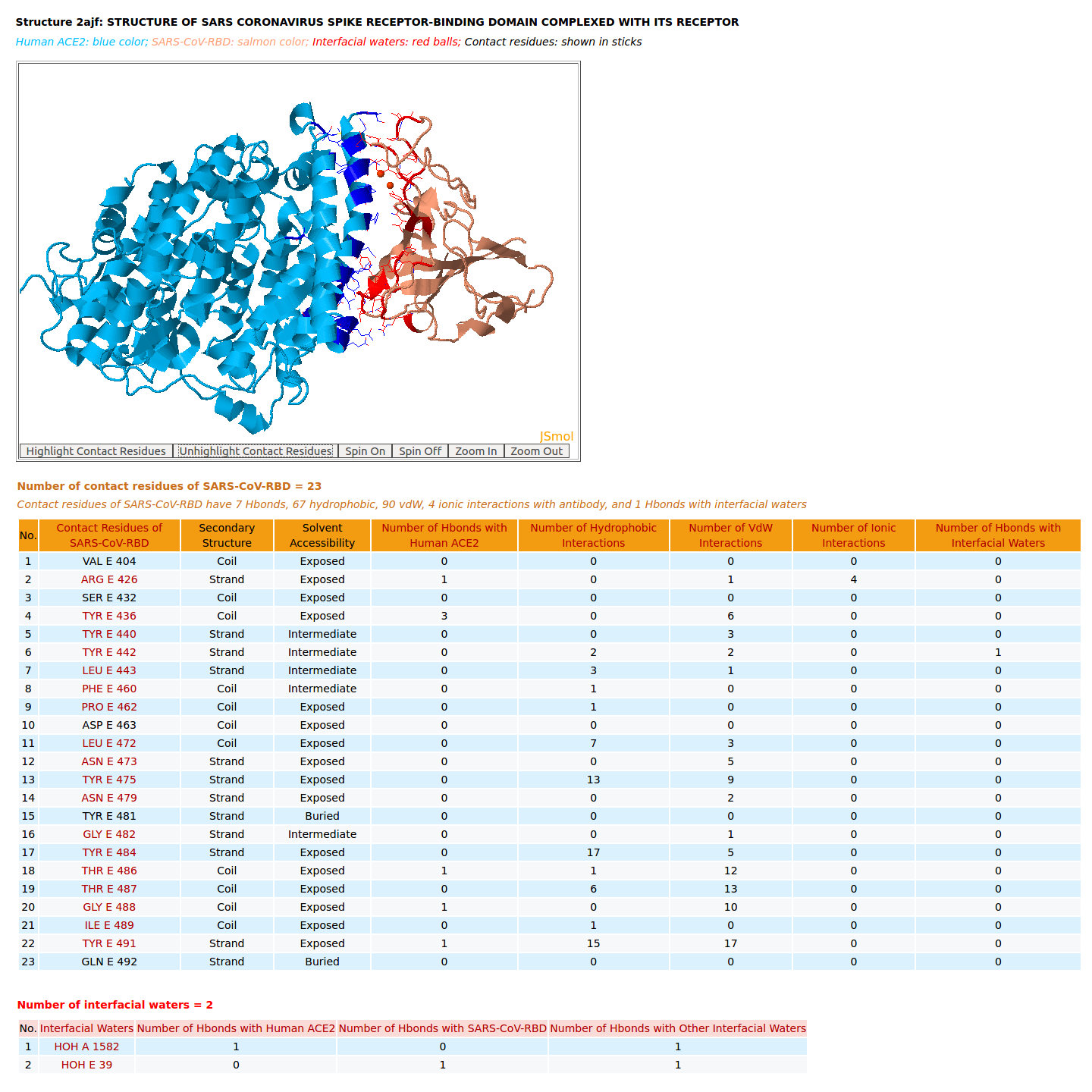
|
|
AppA displays the rendition of 3D structure and the table of of binding residues of SARS-CoV-RBD of structure (2AJF) with human ACE2. The binding residues of SARS-CoV-RBD are highlighted in salmon sticks. The number of interactions at the interface of SARS-CoV-RBD and ACE2 in the structure 2AJF are 7 Hbonds, 67 hydrophobic, 90 vdW, 4 ionic interactions, 1 Hbond with interfacial waters. |
[back to top] |
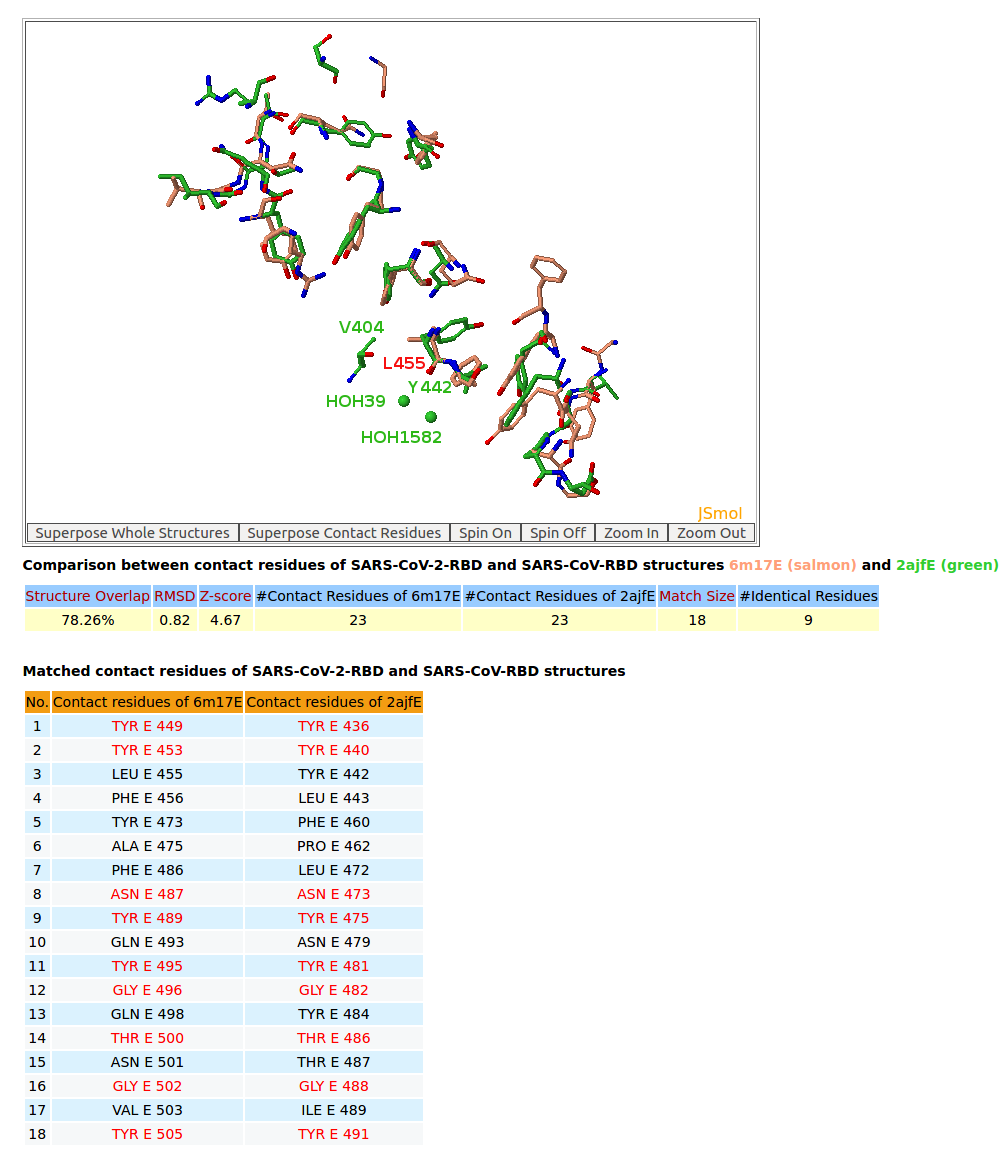
| 5. 3D structure comparison between binding residues of SARS-CoV-2-RBD (6M17) and SARS-CoV-RBD (2AJF) |
|
|
AppA displays 3D structure comparison between the binding residues of SARS-CoV-2-RBD (6M17) and SARS-CoV-RBD (2AJF) with human ACE2. As seen in the table, the conserved binding residues of SARS-CoV-2-RBD (6M17) and SARS-CoV-RBD (2AJF) with human ACE2 are highlighed red. |
[back to top] |
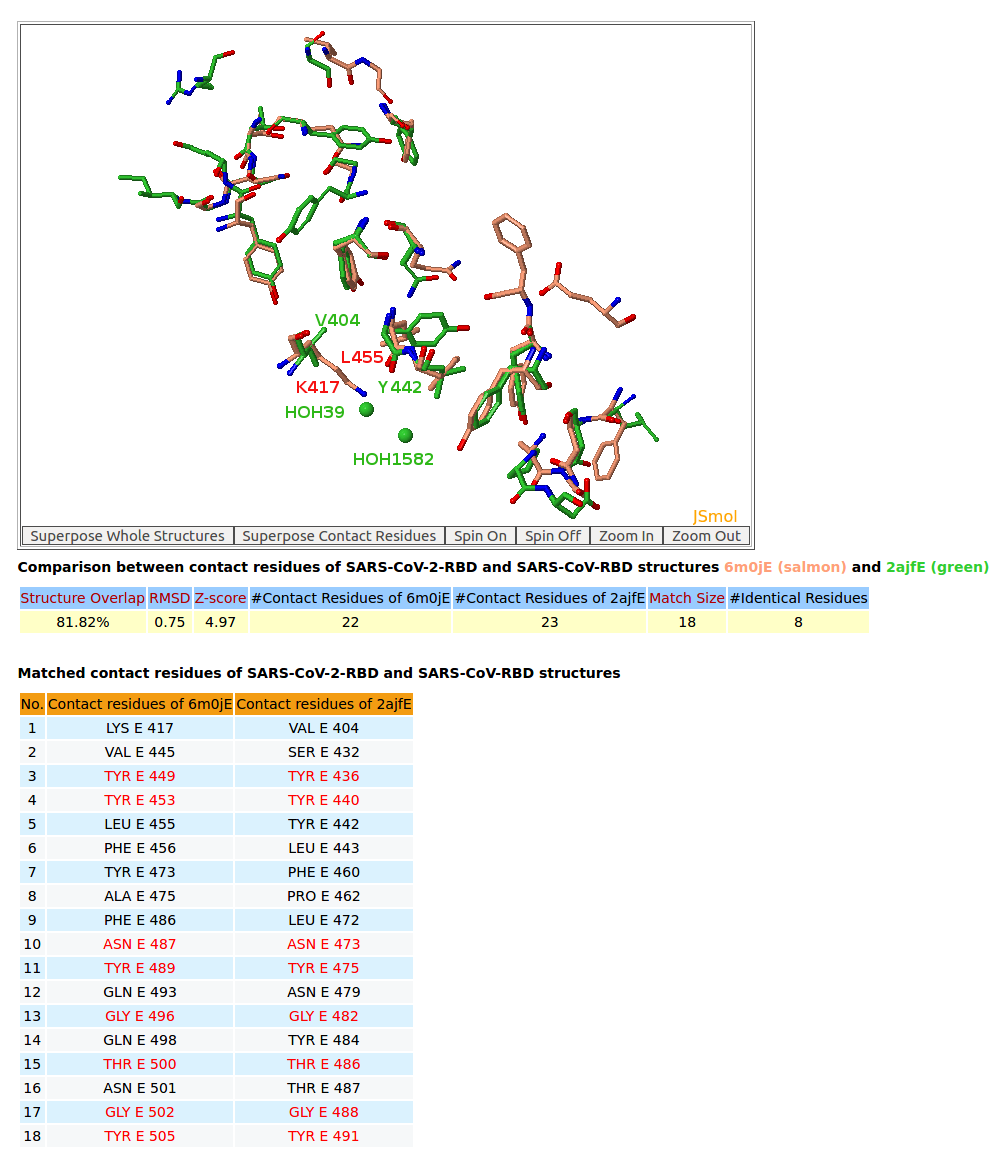
| 6. 3D structure comparison between binding residues of SARS-CoV-2-RBD (6M0J) and SARS-CoV-RBD (2AJF) |
|
|
AppA displays 3D structure comparison between the binding residues of SARS-CoV-2-RBD (6M0J) and SARS-CoV-RBD (2AJF) with human ACE2. The conserved binding residues of SARS-CoV-2-RBD (6M0J) and SARS-CoV-RBD (2AJF) with human ACE2 are highlighed red in the table. |
[back to top] |
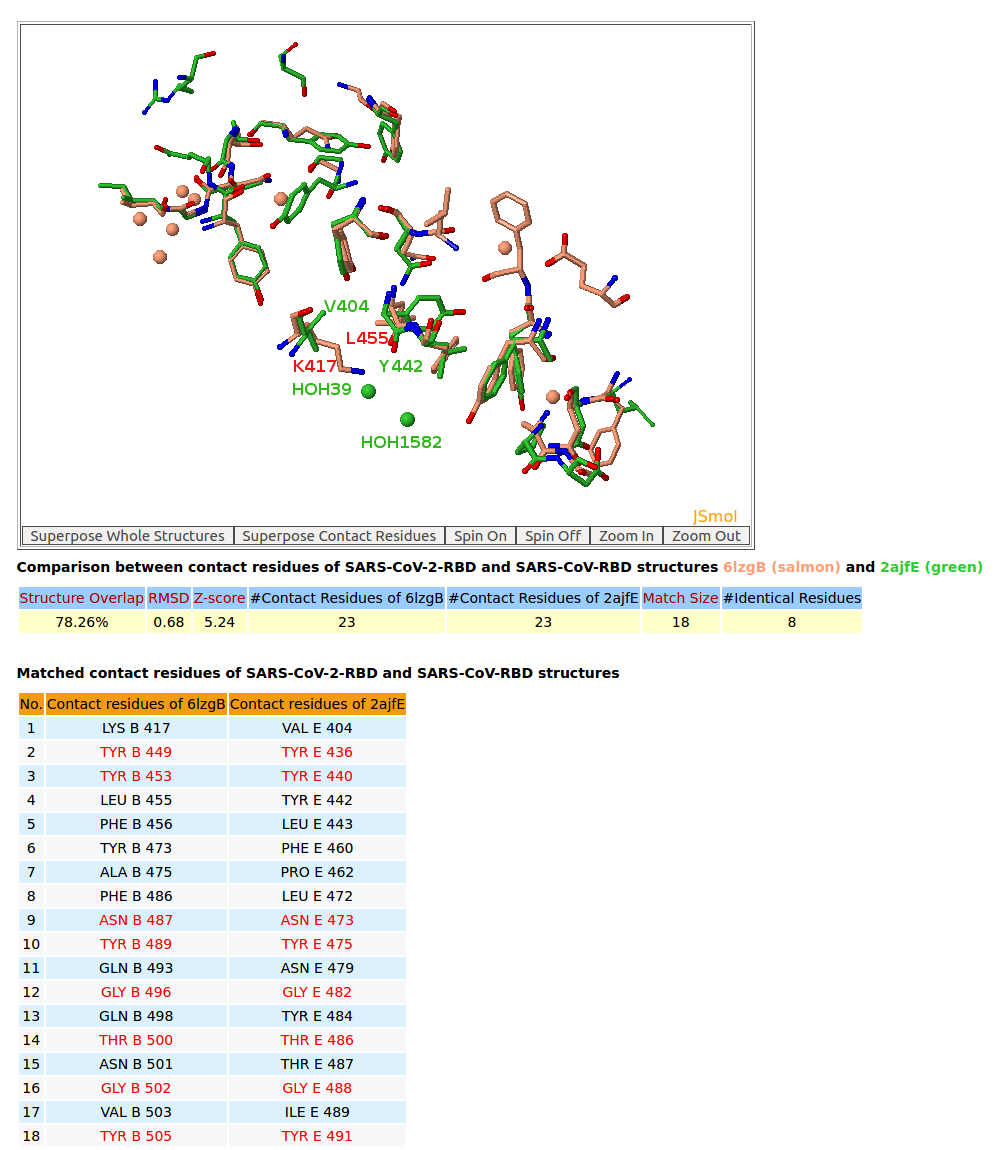
| 7. 3D structure comparison between binding residues of SARS-CoV-2-RBD (6LZG) and SARS-CoV-RBD (2AJF) |
|
|
AppA displays 3D structure comparison between the binding residues of SARS-CoV-2-RBD (6LZG) and SARS-CoV-RBD (2AJF) with human ACE2. The interfaces of SARS-CoV-2-RBD of all three structures 6M17, 6M0J, 6LZG have eight residues Tyr449, Tyr453, Asn487, Tyr489, Gly496, Thr500, Gly502, Tyr505 that are not only conserved but also occupy identical spacial locations with Tyr436, Tyr440, Asn473, Tyr475, Gly482, Thr486, Gly488, Tyr491 of SARS-CoV-RBD (2AJF). Among three structures, 6LZG and 6M0J indicate that these eight binding residues with four conserved Tyr of SARS-CoV-2-RBD have either Hbonds with ACE2 or Hbonds with interfacial waters that significantly support for convergent evolution between SARS-CoV-RBD and SARS-CoV-2-RBD for binding ACE2. It is interesting to note that Tyr also plays an especially important role in interactions between antibody paratopes and antigen epitopes, and the binding residues of SARS-CoV-2-RBD are mainly on three loops like antibody paratopes. In addition, on the interface between SARS-CoV-RBD with ACE2 from the structure 2AJF, two interfacial water molecules (HOH39 and HOH1582) bridge the main chain of Tyr442 of SARS-CoV-RBD and the sidechain of Asp30 of ACE2. From SARS-CoV-RBD to SARS-CoV-2-RBD, the co-substitutions Val404->Lys417 and Tyr442->Leu455 have first broken this interfacial water network as the sidechain of Lys417 is very close to the spatial location of HOH39. After that Lys417 of SARS-CoV-2-RBD makes an important salt bridge with Asp30 of ACE2 as shown in the structures 6M0J and 6LZG that results in improving binding affinity SARS-CoV-2-RBD for ACE2. |
[back to top] |