|
|
|
|

|
|
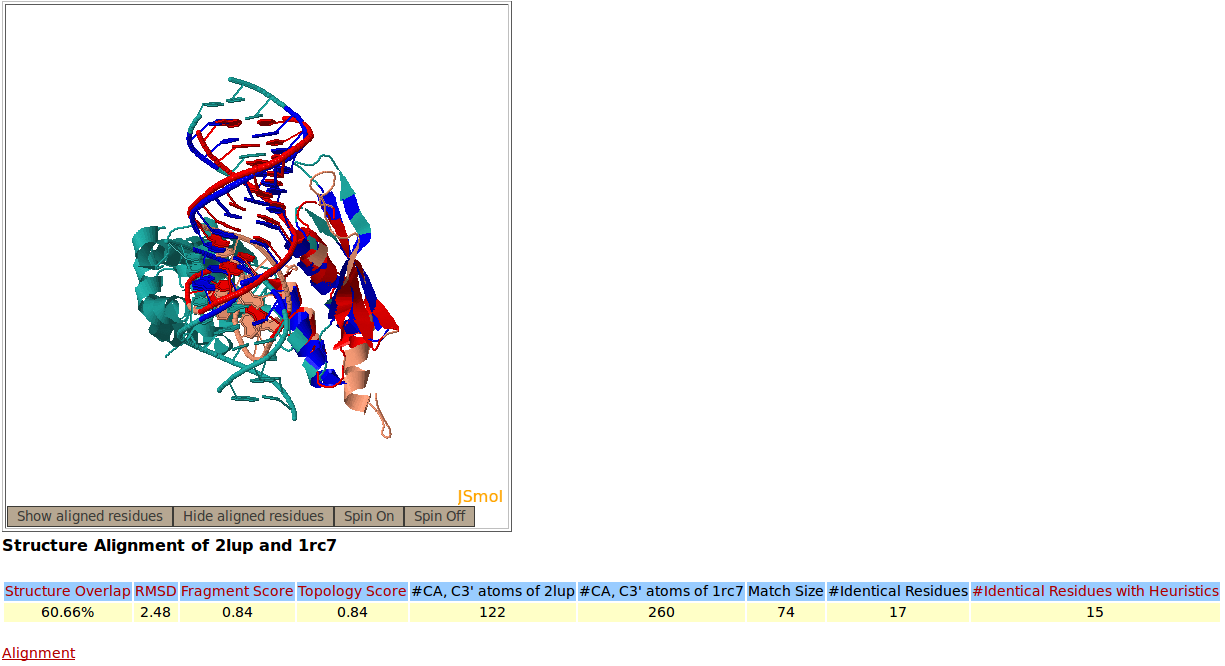
Snapshots of the Rclick server showing the output of a structural alignment of two RNA that belong to Ribonuclease III family, PDB codes 2LUP and 1RC7. Rclick shows the accurate alignment of RNA-protein interactions between 2LUP (salmon) and 1RC7 (green). The superimposed residues of 2LUP and 1RC7 are shown in red and blue colors, respectively. The conserved residues are shown in bold and red lettering.
|
|
| [back to top] |
|
|
|
|

|
|
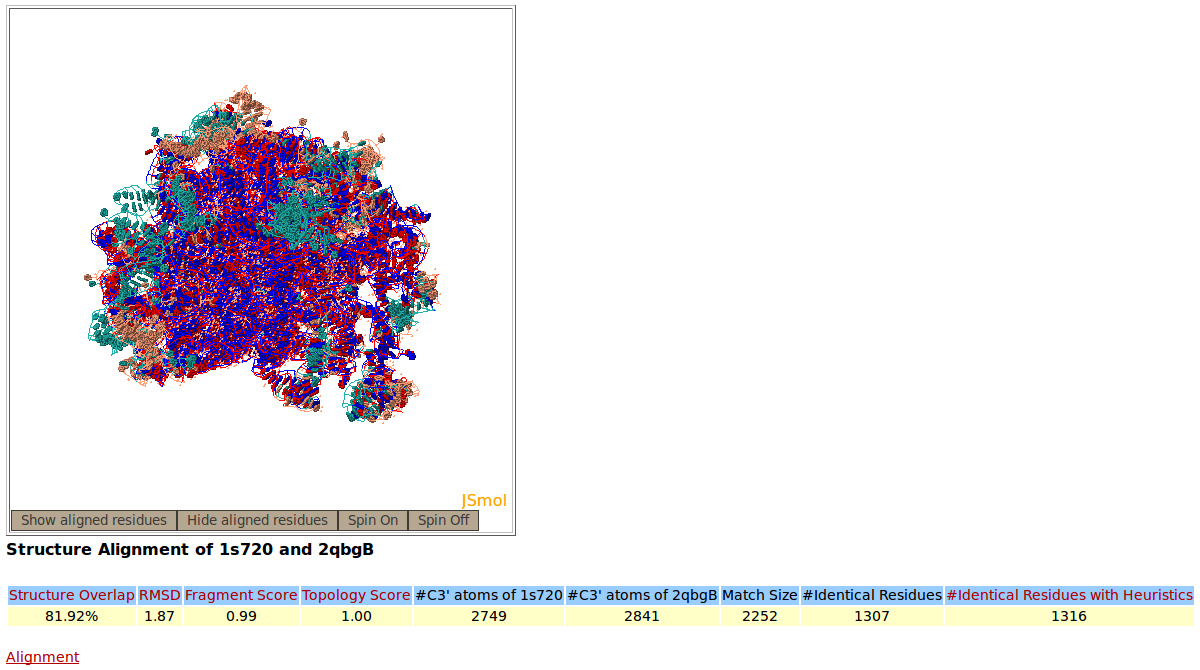
Rclick superimposition of large ribosomal subunit (23S) from Haloarcula marismortui (1S72 chain 0, salmon color) and Escherichia coli (2QBG chain B, green). The superimposed residues of 1S72:0 and 2QBG:B are shown in red and blue, respectively.
|
|
| [back to top] |
|
|
|
|
|
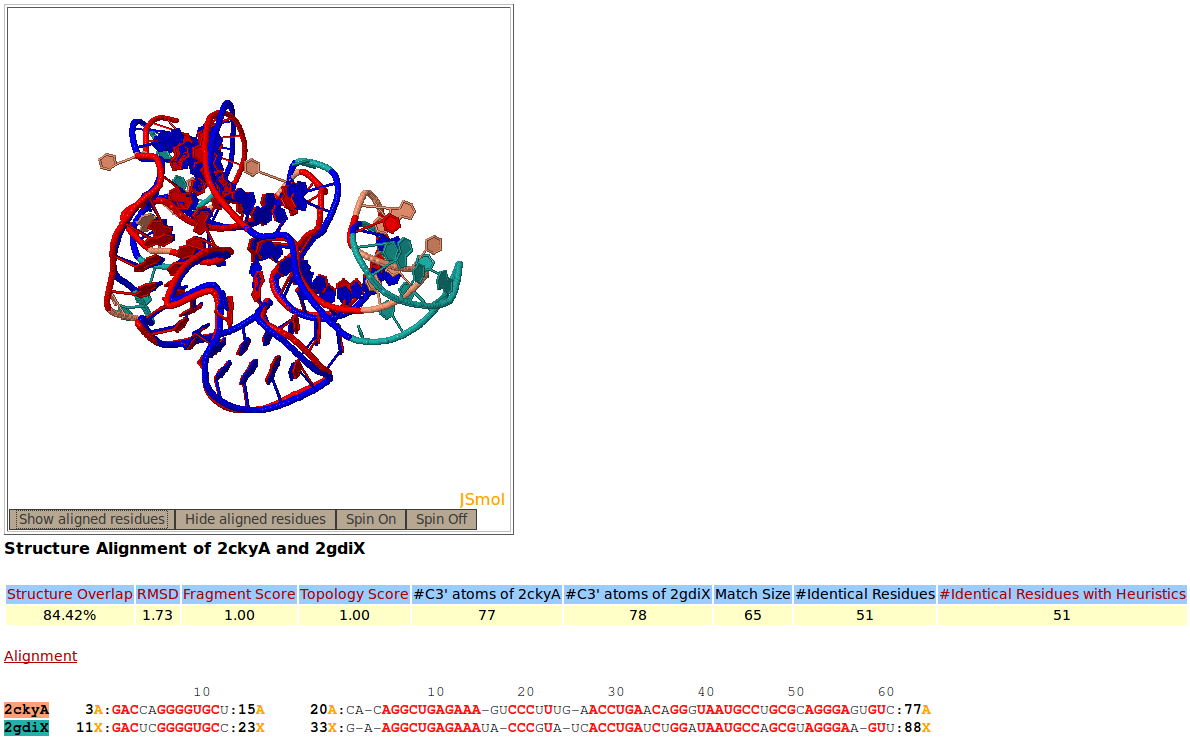
Rclick superimposition of two thiamine pyrophosphate (TPP) riboswitches, PDB codes 2CKY:A (salmon) and 2GDI:X (green). The superimposed residues of 2CKY:A and 2GDI:X are shown in red and blue,respectively.
|
|
|
|
|
|
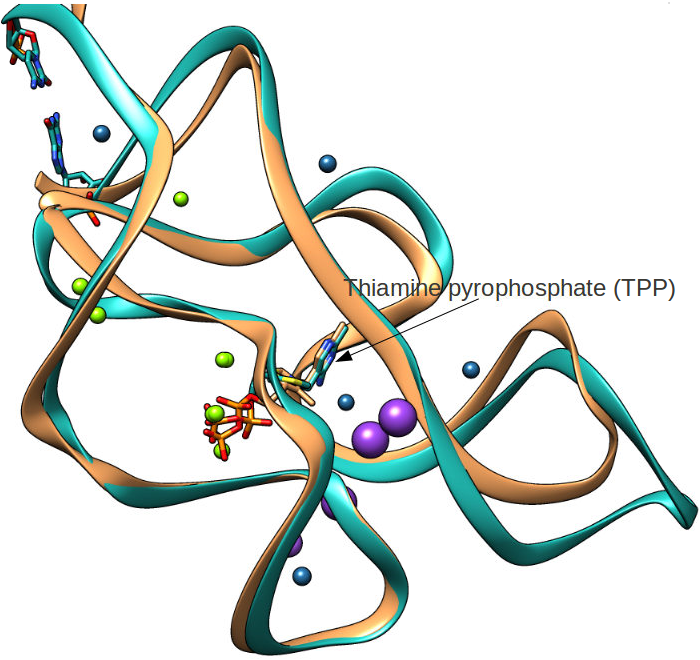
Picture rendered using Chimera (https://www.cgl.ucsf.edu/chimera/)
|
|
| [back to top] |
|
|
|
|
|
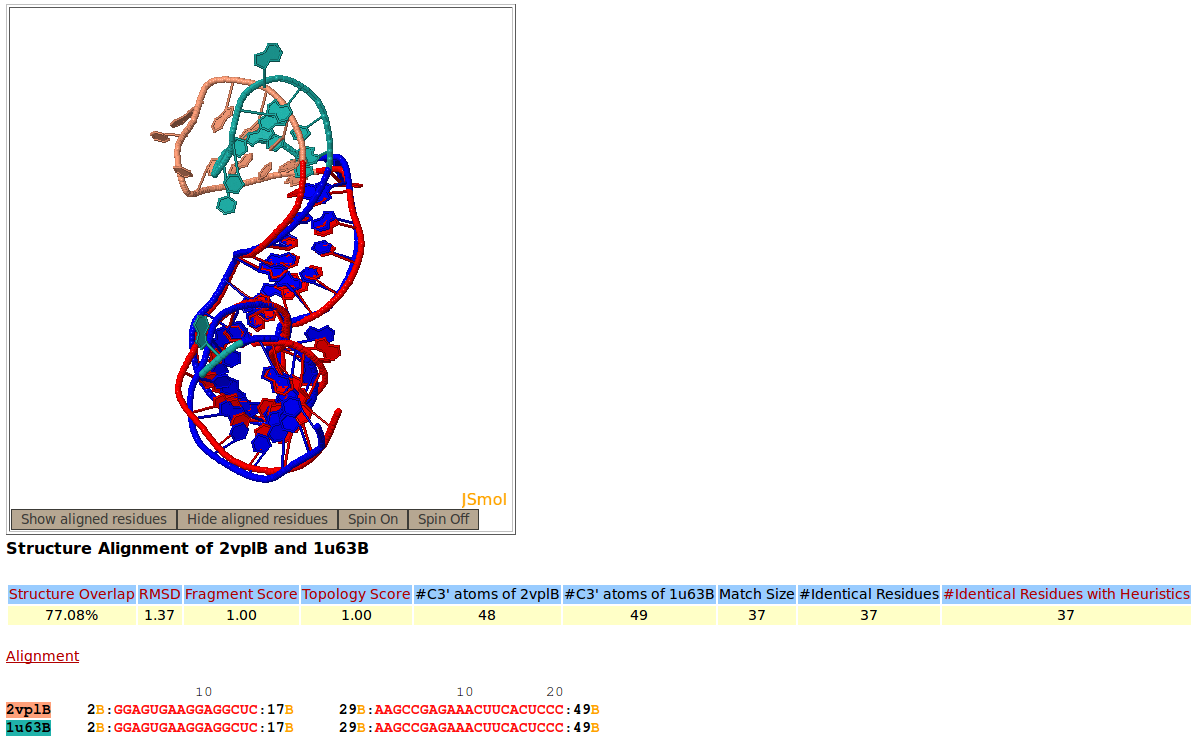
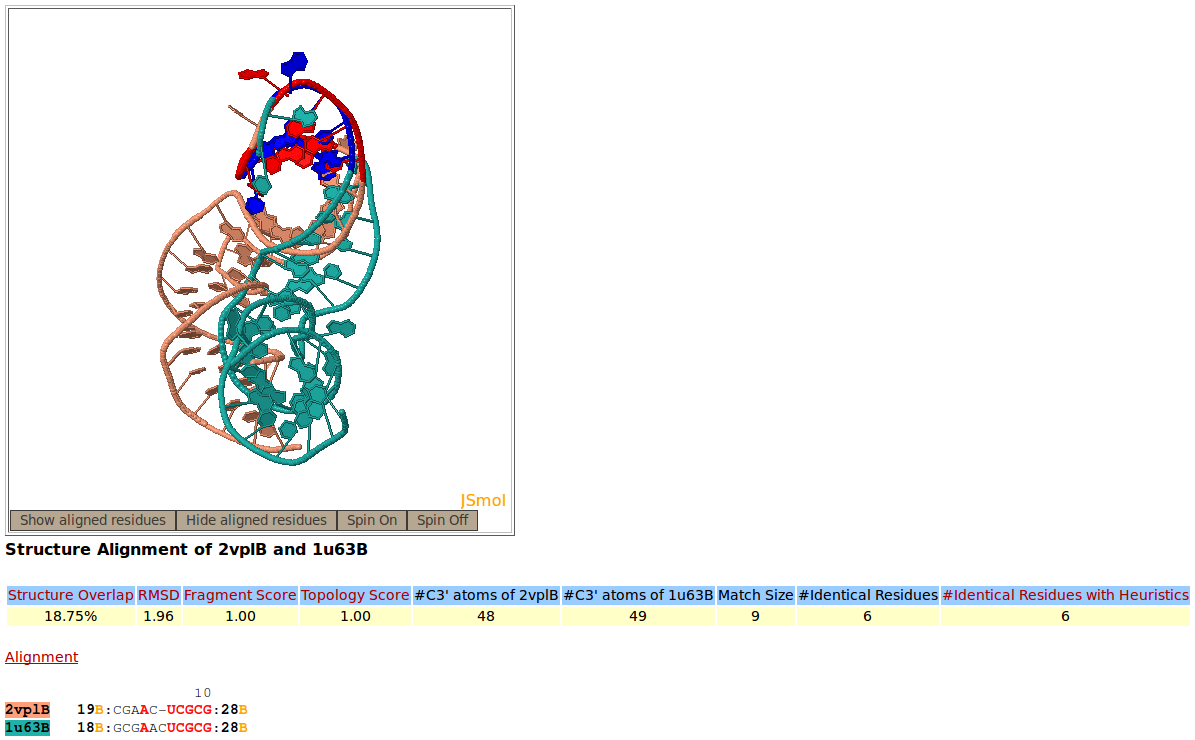
Two structures of fragment of mRNA for ribosomal protein L1, PDB codes 2VPL:B (salmon), and 1U63:B (green), are superimposed against one another by Rclick. The superimposed residues of 2VPL:B and 1U63:B are shown in red and blue,respectively. The regions, spanning residues 2-17 and 29-49 are aligned with one another. Because of a conformational change, the regions of residues 18-28 do not align structurally.
|
|
|
|
|
|
| [back to top] |
|
|
|
|
|
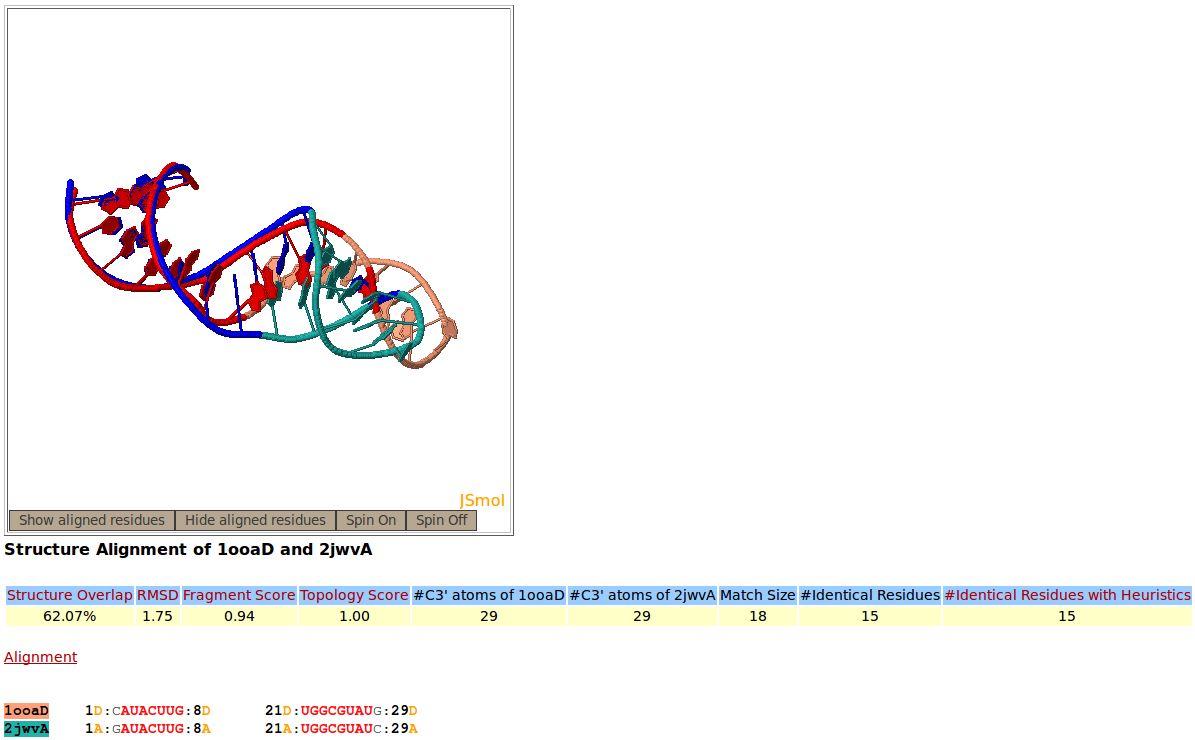
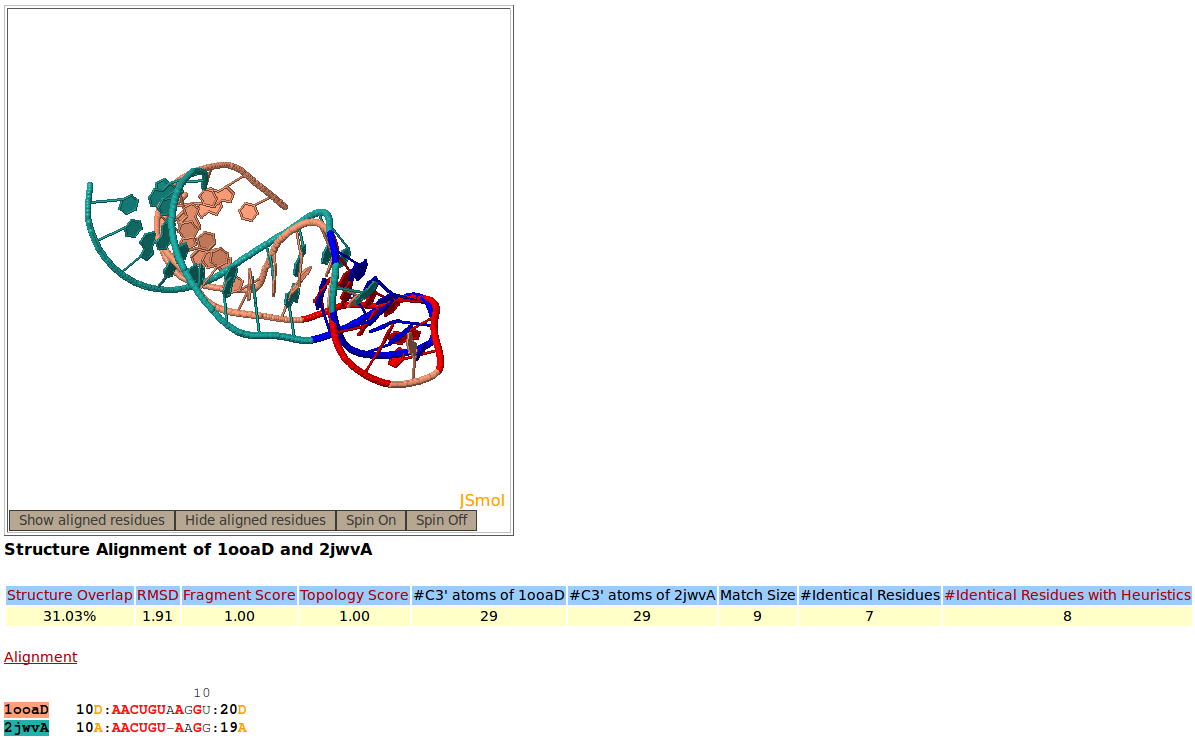
Snapshot of the Rclick showing the first alignment of two RNA aptamer structures, PDB codes 1OOA:D (salmon), and 2JWV:A (green). The superimposed residues of 1OOA:D and 2JWV:A are shown in red and blue,respectively. Because of a conformational change, the regions of residues 10-20 do not align structurally.
|
|
|
|
|
|
| [back to top] |


















